Functional Class I and II Amino Acid-activating Enzymes Can Be Coded by Opposite Strands of the Same Gene
- PMID: 26088142
- PMCID: PMC4528134
- DOI: 10.1074/jbc.M115.642876
Functional Class I and II Amino Acid-activating Enzymes Can Be Coded by Opposite Strands of the Same Gene
Erratum in
-
Functional Class I and II amino acid-activating enzymes can be coded by opposite strands of the same gene.J Biol Chem. 2016 Nov 4;291(45):23830-23831. doi: 10.1074/jbc.A115.642876. J Biol Chem. 2016. PMID: 27815453 Free PMC article. No abstract available.
Abstract
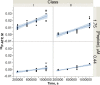
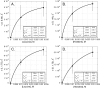
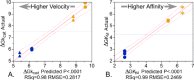
Aminoacyl-tRNA synthetases (aaRS) catalyze both chemical steps that translate the universal genetic code. Rodin and Ohno offered an explanation for the existence of two aaRS classes, observing that codons for the most highly conserved Class I active-site residues are anticodons for corresponding Class II active-site residues. They proposed that the two classes arose simultaneously, by translation of opposite strands from the same gene. We have characterized wild-type 46-residue peptides containing ATP-binding sites of Class I and II synthetases and those coded by a gene designed by Rosetta to encode the corresponding peptides on opposite strands. Catalysis by WT and designed peptides is saturable, and the designed peptides are sensitive to active-site residue mutation. All have comparable apparent second-order rate constants 2.9-7.0E-3 M(-1) s(-1) or ∼750,000-1,300,000 times the uncatalyzed rate. The activities of the two complementary peptides demonstrate that the unique information in a gene can have two functional interpretations, one from each complementary strand. The peptides contain phylogenetic signatures of longer, more sophisticated catalysts we call Urzymes and are short enough to bridge the gap between them and simpler uncoded peptides. Thus, they directly substantiate the sense/antisense coding ancestry of Class I and II aaRS. Furthermore, designed 46-mers achieve similar catalytic proficiency to wild-type 46-mers by significant increases in both kcat and Km values, supporting suggestions that the earliest peptide catalysts activated ATP for biosynthetic purposes.
Keywords: ATP; Rodin-Ohno hypothesis; amino acid activation; aminoacyl tRNA synthetase; chemical biology; enzyme catalysis; gene structure; origin of life; protein design; sense/antisense genetic coding.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures







References
-
- Carter C. W., Jr., Li L., Weinreb V., Collier M., Gonzalez-Rivera K., Jimenez-Rodriguez M., Erdogan O., Kuhlman B., Ambroggio X., Williams T., Chandrasekharan S. N. (2014) The Rodin-Ohno hypothesis that two enzyme superfamilies descended from one ancestral gene: an unlikely scenario for the origins of translation that will not be dismissed. Biol. Direct 9, 11. - PMC - PubMed
-
- Rodin S. N., Ohno S. (1995) Two types of aminoacyl-tRNA synthetases could be originally encoded by complementary strands of the same nucleic acid. Orig. Life Evol. Biosph. 25, 565–589 - PubMed
-
- Pham Y., Li L., Kim A., Erdogan O., Weinreb V., Butterfoss G. L., Kuhlman B., Carter C. W., Jr. (2007) A minimal TrpRS catalytic domain supports sense/antisense ancestry of class I and II aminoacyl-tRNA synthetases. Mol. Cell 25, 851–862 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

