Coxsackievirus can exploit LC3 in both autophagy-dependent and -independent manners in vivo
- PMID: 26090585
- PMCID: PMC4590631
- DOI: 10.1080/15548627.2015.1063769
Coxsackievirus can exploit LC3 in both autophagy-dependent and -independent manners in vivo
Abstract
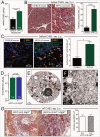
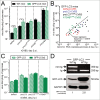
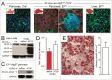
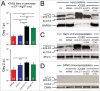
RNA viruses modify intracellular membranes to produce replication scaffolds. In pancreatic cells, coxsackievirus B3 (CVB3) hijacks membranes from the autophagy pathway, and in vivo disruption of acinar cell autophagy dramatically delays CVB3 replication. This is reversed by expression of GFP-LC3, indicating that CVB3 may acquire membranes from an alternative, autophagy-independent, source(s). Herein, using 3 recombinant CVB3s (rCVB3s) encoding different proteins (proLC3, proLC3(G120A), or ATG4B(C74A)), we show that CVB3 is, indeed, flexible in its utilization of cellular membranes. When compared with a control rCVB3, all 3 viruses replicated to high titers in vivo, and caused severe pancreatitis. Most importantly, each virus appeared to subvert membranes in a unique manner. The proLC3 virus produced a large quantity of LC3-I which binds to phosphatidylethanolamine (PE), affording access to the autophagy pathway. The proLC3(G120A) protein cannot attach to PE, and instead binds to the ER-resident protein SEL1L, potentially providing an autophagy-independent source of membranes. Finally, the ATG4B(C74A) protein sequestered host cell LC3-I, causing accumulation of immature phagophores, and massive membrane rearrangement. Taken together, our data indicate that some RNA viruses can exploit a variety of different intracellular membranes, potentially maximizing their replication in each of the diverse cell types that they infect in vivo.
Keywords: ER; ERAD; LC3; RNA virus; SEL1L; autophagy; coxsackievirus; enterovirus; membranes; pancreas.
Figures



References
-
- Uetz P, Dong YA, Zeretzke C, Atzler C, Baiker A, Berger B, Rajagopala SV, Roupelieva M, Rose D, Fossum E, et al.. Herpesviral protein networks and their interaction with the human proteome. Science 2006; 311:239-42; PMID:16339411; http://dx.doi.org/10.1126/science.1116804 - DOI - PubMed
-
- Miller S, Krijnse-Locker J. Modification of intracellular membrane structures for virus replication. Nat Rev Microbiol 2008; 6:363-74; PMID:18414501; http://dx.doi.org/10.1038/nrmicro1890 - DOI - PMC - PubMed
-
- Kirkegaard K. Subversion of the cellular autophagy pathway by viruses. Curr Top Microbiol Immunol 2009; 335:323-33; PMID:19802573 - PubMed
-
- Dales S, Eggers HJ, Tamm I, Palade GE. Electron microscopic study of the formation of poliovirus. Virology 1965; 26:379-89; PMID:14319710; http://dx.doi.org/10.1016/0042-6822(65)90001-2 - DOI - PubMed
-
- Jezequel AM, Steiner JW. Some ultrastructural and histochemical aspects of Coxsackie virus-cell interactions. Lab Invest 1966; 15:1055-83; PMID:5911943 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
