Phylogeny and physiology of candidate phylum 'Atribacteria' (OP9/JS1) inferred from cultivation-independent genomics
- PMID: 26090992
- PMCID: PMC4737943
- DOI: 10.1038/ismej.2015.97
Phylogeny and physiology of candidate phylum 'Atribacteria' (OP9/JS1) inferred from cultivation-independent genomics
Abstract
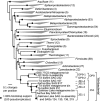
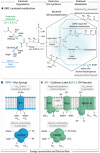
The 'Atribacteria' is a candidate phylum in the Bacteria recently proposed to include members of the OP9 and JS1 lineages. OP9 and JS1 are globally distributed, and in some cases abundant, in anaerobic marine sediments, geothermal environments, anaerobic digesters and reactors and petroleum reservoirs. However, the monophyly of OP9 and JS1 has been questioned and their physiology and ecology remain largely enigmatic due to a lack of cultivated representatives. Here cultivation-independent genomic approaches were used to provide a first comprehensive view of the phylogeny, conserved genomic features and metabolic potential of members of this ubiquitous candidate phylum. Previously available and heretofore unpublished OP9 and JS1 single-cell genomic data sets were used as recruitment platforms for the reconstruction of atribacterial metagenome bins from a terephthalate-degrading reactor biofilm and from the monimolimnion of meromictic Sakinaw Lake. The single-cell genomes and metagenome bins together comprise six species- to genus-level groups that represent most major lineages within OP9 and JS1. Phylogenomic analyses of these combined data sets confirmed the monophyly of the 'Atribacteria' inclusive of OP9 and JS1. Additional conserved features within the 'Atribacteria' were identified, including a gene cluster encoding putative bacterial microcompartments that may be involved in aldehyde and sugar metabolism, energy conservation and carbon storage. Comparative analysis of the metabolic potential inferred from these data sets revealed that members of the 'Atribacteria' are likely to be heterotrophic anaerobes that lack respiratory capacity, with some lineages predicted to specialize in either primary fermentation of carbohydrates or secondary fermentation of organic acids, such as propionate.
Figures


References
-
- Baker BJ, Dick GJ. (2013). Omic approaches in microbial ecology: charting the unknown. Microbe 8: 353–360.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

