Cobalt(III) Protoporphyrin Activates the DGCR8 Protein and Can Compensate microRNA Processing Deficiency
- PMID: 26091172
- PMCID: PMC4788496
- DOI: 10.1016/j.chembiol.2015.05.015
Cobalt(III) Protoporphyrin Activates the DGCR8 Protein and Can Compensate microRNA Processing Deficiency
Abstract
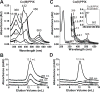
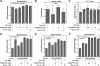
Processing of microRNA primary transcripts (pri-miRNAs) is highly regulated and defects in the processing machinery play a key role in many human diseases. In 22q11.2 deletion syndrome (22q11.2DS), heterozygous deletion of DiGeorge critical region gene 8 (DGCR8) causes a processing deficiency, which contributes to abnormal brain development. The DGCR8 protein is the RNA-binding partner of Drosha RNase, both essential for processing canonical pri-miRNAs. To identify an agent that can compensate reduced DGCR8 expression, we screened for metalloporphyrins that can mimic the natural DGCR8 heme cofactor. We found that Co(III) protoporphyrin IX (PPIX) stably binds DGCR8 and activates it for pri-miRNA processing in vitro and in HeLa cells. Importantly, treating cultured Dgcr8(+/-) mouse neurons with Co(III)PPIX can compensate the pri-miRNA processing defects. Co(III)PPIX is effective at concentrations as low as 0.2 μM and is not degraded by heme degradation enzymes, making it useful as a research tool and a potential therapeutic.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures

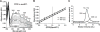
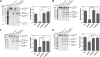

References
-
- Beveridge NJ, Cairns MJ. MicroRNA dysregulation in schizophrenia. Neurobiol Dis. 2012;46:263–271. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

