Efficient Detection of Pathogenic Leptospires Using 16S Ribosomal RNA
- PMID: 26091292
- PMCID: PMC4474562
- DOI: 10.1371/journal.pone.0128913
Efficient Detection of Pathogenic Leptospires Using 16S Ribosomal RNA
Abstract
Pathogenic Leptospira species cause a prevalent yet neglected zoonotic disease with mild to life-threatening complications in a variety of susceptible animals and humans. Diagnosis of leptospirosis, which primarily relies on antiquated serotyping methods, is particularly challenging due to presentation of non-specific symptoms shared by other febrile illnesses, often leading to misdiagnosis. Initiation of antimicrobial therapy during early infection to prevent more serious complications of disseminated infection is often not performed because of a lack of efficient diagnostic tests. Here we report that specific regions of leptospiral 16S ribosomal RNA molecules constitute a novel and efficient diagnostic target for PCR-based detection of pathogenic Leptospira serovars. Our diagnostic test using spiked human blood was at least 100-fold more sensitive than corresponding leptospiral DNA-based quantitative PCR assays, targeting the same 16S nucleotide sequence in the RNA and DNA molecules. The sensitivity and specificity of our RNA assay against laboratory-confirmed human leptospirosis clinical samples were 64% and 100%, respectively, which was superior then an established parallel DNA detection assay. Remarkably, we discovered that 16S transcripts remain appreciably stable ex vivo, including untreated and stored human blood samples, further highlighting their use for clinical detection of L. interrogans. Together, these studies underscore a novel utility of RNA targets, specifically 16S rRNA, for development of PCR-based modalities for diagnosis of human leptospirosis, and also may serve as paradigm for detection of additional bacterial pathogens for which early diagnosis is warranted.
Conflict of interest statement
Figures
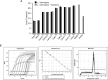


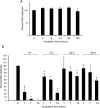
References
-
- Bharti AR, Nally JE, Ricaldi JN, Matthias MA, Diaz MM, Lovett MA, et al. Leptospirosis: a zoonotic disease of global importance. Lancet Infect Dis. 2003;3(12):757–71. . - PubMed
-
- Ko AI, Galvao Reis M, Ribeiro Dourado CM, Johnson WD Jr, Riley LW. Urban epidemic of severe leptospirosis in Brazil. Salvador Leptospirosis Study Group. Lancet. 1999;354(9181):820–5. . - PubMed
-
- McBride AJ, Athanazio DA, Reis MG, Ko AI. Leptospirosis. Curr Opin Infect Dis. 2005;18(5):376–86. . - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

