The Promise of Proteomics for the Study of ADP-Ribosylation
- PMID: 26091340
- PMCID: PMC4486045
- DOI: 10.1016/j.molcel.2015.06.012
The Promise of Proteomics for the Study of ADP-Ribosylation
Abstract
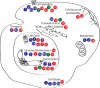
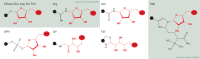
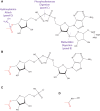
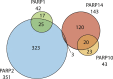
ADP-ribosylation is a post-translational modification where single units (mono-ADP-ribosylation) or polymeric chains (poly-ADP-ribosylation) of ADP-ribose are conjugated to proteins by ADP-ribosyltransferases. This post-translational modification and the ADP-ribosyltransferases (also known as PARPs) responsible for its synthesis have been found to play a role in nearly all major cellular processes, including DNA repair, transcription, translation, cell signaling, and cell death. Furthermore, dysregulation of ADP-ribosylation has been linked to diseases including cancers, diabetes, neurodegenerative disorders, and heart failure, leading to the development of therapeutic PARP inhibitors, many of which are currently in clinical trials. The study of this therapeutically important modification has recently been bolstered by the application of mass spectrometry-based proteomics, arguably the most powerful tool for the unbiased analysis of protein modifications. Unfortunately, progress has been hampered by the inherent challenges that stem from the physicochemical properties of ADP-ribose, which as a post-translational modification is highly charged, heterogeneous (linear or branched polymers, as well as monomers), labile, and found on a wide range of amino acid acceptors. In this Perspective, we discuss the progress that has been made in addressing these challenges, including the recent breakthroughs in proteomics techniques to identify ADP-ribosylation sites, and future developments to provide a proteome-wide view of the many cellular processes regulated by ADP-ribosylation.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures


References
-
- Adamietz P., Hilz H. Poly(adenosine diphosphate ribose) is covalently linked to nuclear proteins by two types of bonds. Hoppe Seylers Z. Physiol. Chem. 1976;357:527–534. - PubMed
-
- Ahel I., Ahel D., Matsusaka T., Clark A.J., Pines J., Boulton S.J., West S.C. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008;451:81–85. - PubMed
-
- Alvarez-Gonzalez R., Jacobson M.K. Characterization of polymers of adenosine diphosphate ribose generated in vitro and in vivo. Biochemistry. 1987;26:3218–3224. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

