Population structure and history of the Welsh sheep breeds determined by whole genome genotyping
- PMID: 26091804
- PMCID: PMC4474581
- DOI: 10.1186/s12863-015-0216-x
Population structure and history of the Welsh sheep breeds determined by whole genome genotyping
Abstract
Background: One of the most economically important areas within the Welsh agricultural sector is sheep farming, contributing around £230 million to the UK economy annually. Phenotypic selection over several centuries has generated a number of native sheep breeds, which are presumably adapted to the diverse and challenging landscape of Wales. Little is known about the history, genetic diversity and relationships of these breeds with other European breeds. We genotyped 353 individuals from 18 native Welsh sheep breeds using the Illumina OvineSNP50 array and characterised the genetic structure of these breeds. Our genotyping data were then combined with, and compared to, those from a set of 74 worldwide breeds, previously collected during the International Sheep Genome Consortium HapMap project.
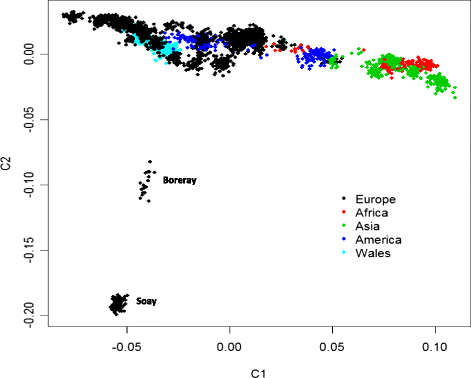
Results: Model based clustering of the Welsh and European breeds indicated shared ancestry. This finding was supported by multidimensional scaling analysis (MDS), which revealed separation of the European, African and Asian breeds. As expected, the commercial Texel and Merino breeds appeared to have extensive co-ancestry with most European breeds. Consistently high levels of haplotype sharing were observed between native Welsh and other European breeds. The Welsh breeds did not, however, form a genetically homogeneous group, with pairwise F ST between breeds averaging 0.107 and ranging between 0.020 and 0.201. Four subpopulations were identified within the 18 native breeds, with high homogeneity observed amongst the majority of mountain breeds. Recent effective population sizes estimated from linkage disequilibrium ranged from 88 to 825.
Conclusions: Welsh breeds are highly diverse with low to moderate effective population sizes and form at least four distinct genetic groups. Our data suggest common ancestry between the native Welsh and European breeds. These findings provide the basis for future genome-wide association studies and a first step towards developing genomics assisted breeding strategies in the UK.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

