MouseMine: a new data warehouse for MGI
- PMID: 26092688
- PMCID: PMC4534495
- DOI: 10.1007/s00335-015-9573-z
MouseMine: a new data warehouse for MGI
Abstract
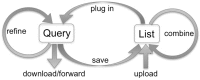
MouseMine (www.mousemine.org) is a new data warehouse for accessing mouse data from Mouse Genome Informatics (MGI). Based on the InterMine software framework, MouseMine supports powerful query, reporting, and analysis capabilities, the ability to save and combine results from different queries, easy integration into larger workflows, and a comprehensive Web Services layer. Through MouseMine, users can access a significant portion of MGI data in new and useful ways. Importantly, MouseMine is also a member of a growing community of online data resources based on InterMine, including those established by other model organism databases. Adopting common interfaces and collaborating on data representation standards are critical to fostering cross-species data analysis. This paper presents a general introduction to MouseMine, presents examples of its use, and discusses the potential for further integration into the MGI interface.
Figures




References
-
- Lyne R, Smith R, Rutherford K, Wakeling M, Varley A, Guillier F, Janssens H, Ji W, Mclaren P, North P, Rana D, Riley T, Sullivan J, Watkins X, Woodbridge M, Lilley K, Russell S, Ashburner M, Mizuguchi K, Micklem G. FlyMine: an integrated database for Drosophila and Anopheles genomics. Genome Biol. 2007;8(7):R129. doi: 10.1186/gb-2007-8-7-r129. - DOI - PMC - PubMed
-
- Smith RN, Aleksic J, Butano D, Carr A, Contrino S, Hu F, Lyne M, Lyne R, Kalderimis A, Rutherford K, Stepan R, Sullivan J, Wakeling M, Watkins X, Micklem G. InterMine: a flexible data warehouse system for the integration and analysis of heterogeneous biological data. Bioinformatics. 2012;28(23):3163–3165. doi: 10.1093/bioinformatics/bts577. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

