MS2 coat proteins bound to yeast mRNAs block 5' to 3' degradation and trap mRNA decay products: implications for the localization of mRNAs by MS2-MCP system
- PMID: 26092944
- PMCID: PMC4509929
- DOI: 10.1261/rna.051797.115
MS2 coat proteins bound to yeast mRNAs block 5' to 3' degradation and trap mRNA decay products: implications for the localization of mRNAs by MS2-MCP system
Abstract
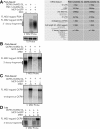
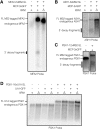
Arrays of MS2 binding sites are placed into mRNAs and are commonly used to visualize the localization of mRNAs in vivo by the expression of an MS2-GFP fusion protein. In Saccharomyces cerevisiae, we observed that arrays of MS2 binding sites inhibit 5' to 3' degradation of the mRNA in yeast cells and lead to the accumulation of a 3' mRNA fragment containing the MS2 binding sites. This accumulation can be dependent on the binding of the MS2 stem loops (MS2-SL) by MS2 coat proteins (MCPs). Since such decay fragments can still bind MCP-GFP, the localization of such mRNA fragments can complicate the interpretation of the localization of full-length mRNA in vivo.
Keywords: MS2-MCP system; mRNA decay; mRNA localization.
© 2015 Garcia and Parker; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures
References
-
- Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. 2005. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 122: 553–563. - PubMed
-
- Haim L, Zipor G, Aronov S, Gerst JE. 2007. A genomic integration method to visualize localization of endogenous mRNAs in living yeast. Nat Methods 4: 409–412. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
