Conceptual and methodological advances in cell-free directed evolution
- PMID: 26093059
- PMCID: PMC4641812
- DOI: 10.1016/j.sbi.2015.04.008
Conceptual and methodological advances in cell-free directed evolution
Abstract
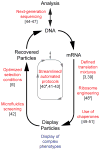
Although cell-free directed evolution methods have been used to engineer proteins for nearly two decades, selections on more complex phenotypes have largely remained in the domain of cell-based engineering approaches. Here, we review recent conceptual advances that now enable in vitro display of multimeric proteins, integral membrane proteins, and proteins with an expanded amino acid repertoire. Additionally, we discuss methodological improvements that have enhanced the accessibility, efficiency, and robustness of cell-free approaches. Coupling these advances with the in vitro advantages of creating exceptionally large libraries and precisely controlling all experimental conditions, cell-free directed evolution is poised to contribute significantly to our understanding and engineering of more complex protein phenotypes.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

