Chimira: analysis of small RNA sequencing data and microRNA modifications
- PMID: 26093149
- PMCID: PMC4595902
- DOI: 10.1093/bioinformatics/btv380
Chimira: analysis of small RNA sequencing data and microRNA modifications
Abstract
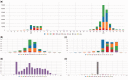
Chimira is a web-based system for microRNA (miRNA) analysis from small RNA-Seq data. Sequences are automatically cleaned, trimmed, size selected and mapped directly to miRNA hairpin sequences. This generates count-based miRNA expression data for subsequent statistical analysis. Moreover, it is capable of identifying epi-transcriptomic modifications in the input sequences. Supported modification types include multiple types of 3'-modifications (e.g. uridylation, adenylation), 5'-modifications and also internal modifications or variation (ADAR editing or single nucleotide polymorphisms). Besides cleaning and mapping of input sequences to miRNAs, Chimira provides a simple and intuitive set of tools for the analysis and interpretation of the results (see also Supplementary Material). These allow the visual study of the differential expression between two specific samples or sets of samples, the identification of the most highly expressed miRNAs within sample pairs (or sets of samples) and also the projection of the modification profile for specific miRNAs across all samples. Other tools have already been published in the past for various types of small RNA-Seq analysis, such as UEA workbench, seqBuster, MAGI, OASIS and CAP-miRSeq, CPSS for modifications identification. A comprehensive comparison of Chimira with each of these tools is provided in the Supplementary Material. Chimira outperforms all of these tools in total execution speed and aims to facilitate simple, fast and reliable analysis of small RNA-Seq data allowing also, for the first time, identification of global microRNA modification profiles in a simple intuitive interface.
Availability and implementation: Chimira has been developed as a web application and it is accessible here: http://www.ebi.ac.uk/research/enright/software/chimira.
Contact: aje@ebi.ac.uk
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author 2015. Published by Oxford University Press.
Figures
References
-
- Heo I., et al. (2009) TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell, 138, 696–708. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

