Light Signaling Mechanism of Two Tandem Bacteriophytochromes
- PMID: 26095026
- PMCID: PMC4497868
- DOI: 10.1016/j.str.2015.04.022
Light Signaling Mechanism of Two Tandem Bacteriophytochromes
Abstract
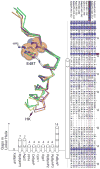
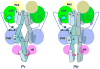
RpBphP2 and RpBphP3, two tandem bacteriophytochromes from the photosynthetic bacterium Rhodopseudomonas palustris, share high sequence identity but exhibit distinct photoconversion behavior. Unlike the canonical RpBphP2, RpBphP3 photoconverts to an unusual near-red-absorbing (Pnr) state; both are required for synthesis of light-harvesting complexes under low-light conditions. Here we report the crystal structures of the photosensory core modules of RpBphP2 and RpBphP3. Despite different quaternary structures, RpBphP2 and RpBphP3 adopt nearly identical tertiary structures. The RpBphP3 structure reveals tongue-and-groove interactions at the interface between the GAF and PHY domains. A single mutation in the PRxSF motif at the GAF-PHY interface abolishes light-induced formation of the Pnr state in RpBphP3, possibly due to altered structural rigidity of the chromophore-binding pocket. Structural comparisons suggest that long-range signaling involves structural rearrangement of the helical spine at the dimer interface. These structures, together with mutational studies, provide insights into photoconversion and the long-range signaling mechanism in phytochromes.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures




References
-
- Bellini D, Papiz MZ. Structure of a Bacteriophytochrome and Light-Stimulated Protomer Swapping with a Gene Repressor. Structure. 2012a;20:1436–1446. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

