Differential Requirements for eIF4E Dose in Normal Development and Cancer
- PMID: 26095252
- PMCID: PMC4491046
- DOI: 10.1016/j.cell.2015.05.049
Differential Requirements for eIF4E Dose in Normal Development and Cancer
Abstract
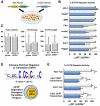
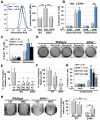
eIF4E, the major cap-binding protein, has long been considered limiting for translating the mammalian genome. However, the eIF4E dose requirement at an organismal level remains unexplored. By generating an Eif4e haploinsufficient mouse, we found that a 50% reduction in eIF4E expression, while compatible with normal development and global protein synthesis, significantly impeded cellular transformation. Genome-wide translational profiling uncovered a translational program induced by oncogenic transformation and revealed a critical role for the dose of eIF4E, specifically in translating a network of mRNAs enriched for a unique 5' UTR signature. In particular, we demonstrate that the dose of eIF4E is essential for translating mRNAs that regulate reactive oxygen species, fueling transformation and cancer cell survival in vivo. Our findings indicate eIF4E is maintained at levels in excess for normal development that are hijacked by cancer cells to drive a translational program supporting tumorigenesis.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures
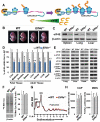
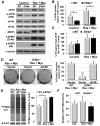
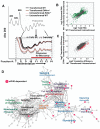
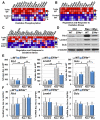

References
-
- Bah A, Vernon RM, Siddiqui Z, Krzeminski M, Muhandiram R, Zhao C, Sonenberg N, Kay LE, Forman-Kay JD. Folding of an intrinsically disordered protein by phosphorylation as a regulatory switch. Nature. 2014 - PubMed
-
- Blackshear PJ, Stumpo DJ, Carballo E, Lawrence JC., Jr. Disruption of the gene encoding the mitogen-regulated translational modulator PHAS-I in mice. The Journal of biological chemistry. 1997;272:31510–31514. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- DP2 OD008509/OD/NIH HHS/United States
- 7DP2OD00850902/OD/NIH HHS/United States
- R01 HD086634/HD/NICHD NIH HHS/United States
- R01 CA184624/CA/NCI NIH HHS/United States
- F32 CA189696/CA/NCI NIH HHS/United States
- R01CA140456/CA/NCI NIH HHS/United States
- P30CA82103/CA/NCI NIH HHS/United States
- R01 CA140456/CA/NCI NIH HHS/United States
- R01 CA154916/CA/NCI NIH HHS/United States
- R01CA184624/CA/NCI NIH HHS/United States
- P30 CA082103/CA/NCI NIH HHS/United States
- R01CA154916/CA/NCI NIH HHS/United States
- 1F32CA189696/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

