GONAD: Genome-editing via Oviductal Nucleic Acids Delivery system: a novel microinjection independent genome engineering method in mice
- PMID: 26096991
- PMCID: PMC4476150
- DOI: 10.1038/srep11406
GONAD: Genome-editing via Oviductal Nucleic Acids Delivery system: a novel microinjection independent genome engineering method in mice
Abstract
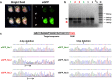
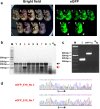
Microinjection is considered the gold standard technique for delivery of nucleic acids (NAs; transgenes or genome editing tools such as CRISPR/Cas9 systems) into embryos, for creating genetically modified organisms. It requires sophisticated equipment as well as well-trained and highly skilled personnel to perform the micro-injection technique. Here, we describe a novel and simple microinjection-independent technique, called Genome-editing via Oviductal Nucleic Acids Delivery (GONAD). Using GONAD, we show that NAs (e.g., eGFP mRNA or Cas9 mRNA/sgRNAs) can be effectively delivered to pre-implantation embryos within the intact mouse oviduct by a simple electroporation method, and result in the desired genetic modification in the embryos. Thus GONAD can bypass many complex steps in transgenic technology such as isolation of zygotes, microinjection of NAs into them, and their subsequent transfer to pseudo-pregnant animals. Furthermore, this method can potentially be used for genome editing in species other than mice.
Figures


References
-
- Sato M., Akasaka E., Saitoh I., Ohtsuka M. & Watanabe S. In vivo gene transfer in mouse preimplantation embryos after intraoviductal injection of plasmid DNA and subsequent in vivo electroporation. Syst Biol Reprod Med 58, 278–287 (2012). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

