The Sequence-Specific Cellular Uptake of Spherical Nucleic Acid Nanoparticle Conjugates
- PMID: 26097111
- PMCID: PMC4560454
- DOI: 10.1002/smll.201500027
The Sequence-Specific Cellular Uptake of Spherical Nucleic Acid Nanoparticle Conjugates
Abstract
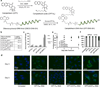
The sequence-dependent cellular uptake of spherical nucleic acid nanoparticle conjugates (SNAs) is investigated. This process occurs by interaction with class A scavenger receptors (SR-A) and caveolae-mediated endocytosis. It is known that linear poly(guanine) (poly G) is a natural ligand for SR-A, and it has been proposed that interaction of poly G with SR-A is dependent on the formation of G-quadruplexes. Since G-rich oligonucleotides are known to interact strongly with SR-A, it is hypothesized that SNAs with higher G contents would be able to enter cells in larger amounts than SNAs composed of other nucleotides, and as such, cellular internalization of SNAs is measured as a function of constituent oligonucleotide sequence. Indeed, SNAs with enriched G content show the highest cellular uptake. Using this hypothesis, a small molecule (camptothecin) is chemically conjugated with SNAs to create drug-SNA conjugates and it is observed that poly G SNAs deliver the most camptothecin to cells and have the highest cytotoxicity in cancer cells. Our data elucidate important design considerations for enhancing the intracellular delivery of spherical nucleic acids.
Keywords: cellular uptake; guanine; nanoparticles; sequence-specific; spherical nucleic acids.
© 2015 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Figures



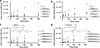
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

