Fresolimumab treatment decreases biomarkers and improves clinical symptoms in systemic sclerosis patients
- PMID: 26098215
- PMCID: PMC4563675
- DOI: 10.1172/JCI77958
Fresolimumab treatment decreases biomarkers and improves clinical symptoms in systemic sclerosis patients
Abstract
Background: TGF-β has potent profibrotic activity in vitro and has long been implicated in systemic sclerosis (SSc), as expression of TGF-β-regulated genes is increased in the skin and lungs of patients with SSc. Therefore, inhibition of TGF-β may benefit these patients.
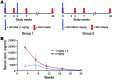
Methods: Patients with early, diffuse cutaneous SSc were enrolled in an open-label trial of fresolimumab, a high-affinity neutralizing antibody that targets all 3 TGF-β isoforms. Seven patients received two 1 mg/kg doses of fresolimumab, and eight patients received one 5 mg/kg dose of fresolimumab. Serial mid-forearm skin biopsies, performed before and after treatment, were analyzed for expression of the TGF-β-regulated biomarker genes thrombospondin-1 (THBS1) and cartilage oligomeric protein (COMP) and stained for myofibroblasts. Clinical skin disease was assessed using the modified Rodnan skin score (MRSS).
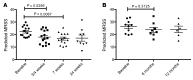
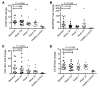
Results: In patient skin, THBS1 expression rapidly declined after fresolimumab treatment in both groups (P = 0.0313 at 7 weeks and P = 0.0156 at 3 weeks), and skin expression of COMP exhibited a strong downward trend in both groups. Clinical skin disease dramatically and rapidly decreased (P < 0.001 at all time points). Expression levels of other TGF-β-regulated genes, including SERPINE1 and CTGF, declined (P = 0.049 and P = 0.012, respectively), and a 2-gene, longitudinal pharmacodynamic biomarker of SSc skin disease decreased after fresolimumab treatment (P = 0.0067). Dermal myofibroblast infiltration also declined in patient skin after fresolimumab (P < 0.05). Baseline levels of THBS1 were predictive of reduced THBS1 expression and improved MRSS after fresolimumab treatment.
Conclusion: The rapid inhibition of TGF-β-regulated gene expression in response to fresolimumab strongly implicates TGF-β in the pathogenesis of fibrosis in SSc. Parallel improvement in the MRSS indicates that fresolimumab rapidly reverses markers of skin fibrosis.
Trial registration: Clinicaltrials.gov NCT01284322.
Figures





References
-
- Derynck R, Miyaono K, eds. The TGF-Beta Family. Cold Spring Harbor Monograph Series 50. Cold Spring Harbor, New York, USA: Cold Spring Harbor Laboratory Press; 2007.
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

