Host Subtraction, Filtering and Assembly Validations for Novel Viral Discovery Using Next Generation Sequencing Data
- PMID: 26098299
- PMCID: PMC4476701
- DOI: 10.1371/journal.pone.0129059
Host Subtraction, Filtering and Assembly Validations for Novel Viral Discovery Using Next Generation Sequencing Data
Abstract
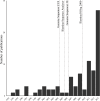
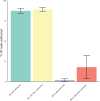
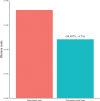
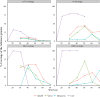
The use of next generation sequencing (NGS) to identify novel viral sequences from eukaryotic tissue samples is challenging. Issues can include the low proportion and copy number of viral reads and the high number of contigs (post-assembly), making subsequent viral analysis difficult. Comparison of assembly algorithms with pre-assembly host-mapping subtraction using a short-read mapping tool, a k-mer frequency based filter and a low complexity filter, has been validated for viral discovery with Illumina data derived from naturally infected liver tissue and simulated data. Assembled contig numbers were significantly reduced (up to 99.97%) by the application of these pre-assembly filtering methods. This approach provides a validated method for maximizing viral contig size as well as reducing the total number of assembled contigs that require down-stream analysis as putative viral nucleic acids.
Conflict of interest statement
Figures




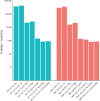
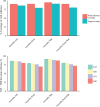
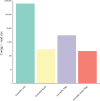


References
-
- Drosten C, Günther S, Preiser W, Van der Werf S, Brodt HR, Becker S et al. (2003) Identification of a novel Coronavirus in patients with severe acute respiratory syndrome N Engl J Med. 348, 1967–1976. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

