Structural Symmetry in Membrane Proteins
- PMID: 26098517
- PMCID: PMC5500171
- DOI: 10.1146/annurev-biophys-051013-023008
Structural Symmetry in Membrane Proteins
Abstract
Symmetry is a common feature among natural systems, including protein structures. A strong propensity toward symmetric architectures has long been recognized for water-soluble proteins, and this propensity has been rationalized from an evolutionary standpoint. Proteins residing in cellular membranes, however, have traditionally been less amenable to structural studies, and thus the prevalence and significance of symmetry in this important class of molecules is not as well understood. In the past two decades, researchers have made great strides in this area, and these advances have provided exciting insights into the range of architectures adopted by membrane proteins. These structural studies have revealed a similarly strong bias toward symmetric arrangements, which were often unexpected and which occurred despite the restrictions imposed by the membrane environment on the possible symmetry groups. Moreover, membrane proteins disproportionately contain internal structural repeats resulting from duplication and fusion of smaller segments. This article discusses the types and origins of symmetry in membrane proteins and the implications of symmetry for protein function.
Keywords: alternating access; asymmetry; internal repeats; inverted-topology repeats; oligomer.
Figures
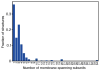


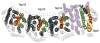
References
-
- Abraham A-L, Pothier J, Rocha EPC. Alternative to Homo-oligomerisation: The Creation of Local Symmetry in Proteins by Internal Amplification. J Mol Biol. 2009;394(3):522–34. - PubMed
-
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408(6814):796–815. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

