Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins
- PMID: 26098576
- PMCID: PMC4612372
- DOI: 10.1038/ncb3192
Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins
Retraction in
-
Retraction Note: Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins.Nat Cell Biol. 2024 Apr;26(4):660. doi: 10.1038/s41556-024-01383-1. Nat Cell Biol. 2024. PMID: 38438803 Free PMC article. No abstract available.
Abstract
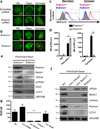
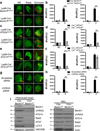
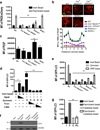
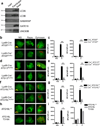
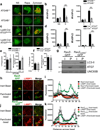
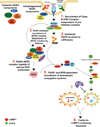
LC3-associated phagocytosis (LAP) is a process wherein elements of autophagy conjugate LC3 to phagosomal membranes. We characterize the molecular requirements for LAP, and identify Rubicon as being required for LAP but not autophagy. Rubicon is recruited to LAPosomes and is required for the activity of a Class III PI(3)K complex containing UVRAG but lacking ATG14 and Ambra1. This allows for the sustained localization of PtdIns(3)P, which is critical for recruitment of downstream autophagic proteins and stabilization of the NOX2 complex to produce reactive oxygen species. Both PtdIns(3)P and reactive oxygen species are required for conjugation of LC3 to LAPosomes and subsequent association with LAMP1(+) lysosomes. LAP is induced by engulfment of Aspergillus fumigatus, a fungal pathogen that commonly afflicts immunocompromised hosts, and is required for its optimal clearance in vivo. Therefore, we have identified molecules that distinguish LAP from canonical autophagy, thereby elucidating the importance of LAP in response to A. fumigatus infection.
Figures


Comment in
-
Rubicon swaps autophagy for LAP.Nat Cell Biol. 2015 Jul;17(7):843-5. doi: 10.1038/ncb3197. Nat Cell Biol. 2015. PMID: 26123110
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

