Oncogenes and tumor suppressor genes: comparative genomics and network perspectives
- PMID: 26099335
- PMCID: PMC4474543
- DOI: 10.1186/1471-2164-16-S7-S8
Oncogenes and tumor suppressor genes: comparative genomics and network perspectives
Abstract
Background: Defective tumor suppressor genes (TSGs) and hyperactive oncogenes (OCGs) heavily contribute to cell proliferation and apoptosis during cancer development through genetic variations such as somatic mutations and deletions. Moreover, they usually do not perform their cellular functions individually but rather execute jointly. Therefore, a comprehensive comparison of their mutation patterns and network properties may provide a deeper understanding of their roles in the cancer development and provide some clues for identification of novel targets.
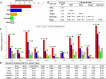
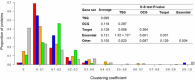
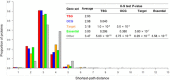
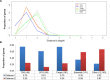
Results: In this study, we performed a comprehensive survey of TSGs and OCGs from the perspectives of somatic mutations and network properties. For comparative purposes, we choose five gene sets: TSGs, OCGs, cancer drug target genes, essential genes, and other genes. Based on the data from Pan-Cancer project, we found that TSGs had the highest mutation frequency in most tumor types and the OCGs second. The essential genes had the lowest mutation frequency in all tumor types. For the network properties in the human protein-protein interaction (PPI) network, we found that, relative to target proteins, essential proteins, and other proteins, the TSG proteins and OCG proteins both tended to have higher degrees, higher betweenness, lower clustering coefficients, and shorter shortest-path distances. Moreover, the TSG proteins and OCG proteins tended to have direct interactions with cancer drug target proteins. To further explore their relationship, we generated a TSG-OCG network and found that TSGs and OCGs connected strongly with each other. The integration of the mutation frequency with the TSG-OCG network offered a network view of TSGs, OCGs, and their interactions, which may provide new insights into how the TSGs and OCGs jointly contribute to the cancer development.
Conclusions: Our study first discovered that the OCGs and TSGs had different mutation patterns, but had similar and stronger protein-protein characteristics relative to the essential proteins or control proteins in the whole human interactome. We also found that the TSGs and OCGs had the most direct interactions with cancer drug targets. The results will be helpful for cancer drug target identification, and ultimately, understanding the etiology of cancer and treatment at the network level.
Figures


References
-
- Society AC. Global cancer facts & figures 2nd edition. American Cancer Society. 2011.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

