Systems analysis of gene ontology and biological pathways involved in post-myocardial infarction responses
- PMID: 26100218
- PMCID: PMC4474415
- DOI: 10.1186/1471-2164-16-S7-S18
Systems analysis of gene ontology and biological pathways involved in post-myocardial infarction responses
Abstract
Background: Pathway analysis has been widely used to gain insight into essential mechanisms of the response to myocardial infarction (MI). Currently, there exist multiple pathway databases that organize molecular datasets and manually curate pathway maps for biological interpretation at varying forms of organization. However, inconsistencies among different databases in pathway descriptions, frequently due to conflicting results in the literature, can generate incorrect interpretations. Furthermore, although pathway analysis software provides detailed images of interactions among molecules, it does not exhibit how pathways interact with one another or with other biological processes under specific conditions.
Methods: We propose a novel method to standardize descriptions of enriched pathways for a set of genes/proteins using Gene Ontology terms. We used this method to examine the relationships among pathways and biological processes for a set of condition-specific genes/proteins, represented as a functional biological pathway-process network. We applied this algorithm to a set of 613 MI-specific proteins we previously identified.
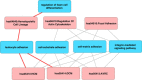
Results: A total of 96 pathways from Biocarta, KEGG, and Reactome, and 448 Gene Ontology Biological Processes were enriched with these 613 proteins. The pathways were represented as Boolean functions of biological processes, delivering an interactive scheme to organize enriched information with an emphasis on involvement of biological processes in pathways. We extracted a network focusing on MI to demonstrate that tyrosine phosphorylation of Signal Transducer and Activator of Transcription (STAT) protein, positive regulation of collagen metabolic process, coagulation, and positive/negative regulation of blood coagulation have immediate impacts on the MI response.
Conclusions: Our method organized biological processes and pathways in an unbiased approach to provide an intuitive way to identify biological properties of pathways under specific conditions. Pathways from different databases have similar descriptions yet diverse biological processes, indicating variation in their ability to share similar functional characteristics. The coverages of pathways can be expanded with the incorporation of more biological processes, predicting involvement of protein members in pathways. Further, detailed analyses of the functional biological pathway-process network will allow researchers and scientists to explore critical routes in biological systems in the progression of disease.
Figures





Similar articles
-
Identification of drug-induced myocardial infarction-related protein targets through the prediction of drug-target interactions and analysis of biological processes.Chem Res Toxicol. 2014 Jul 21;27(7):1263-81. doi: 10.1021/tx500147d. Epub 2014 Jun 18. Chem Res Toxicol. 2014. PMID: 24920530
-
Event ontology: a pathway-centric ontology for biological processes.Pac Symp Biocomput. 2006:152-63. Pac Symp Biocomput. 2006. PMID: 17094236
-
The Pathway Coexpression Network: Revealing pathway relationships.PLoS Comput Biol. 2018 Mar 19;14(3):e1006042. doi: 10.1371/journal.pcbi.1006042. eCollection 2018 Mar. PLoS Comput Biol. 2018. PMID: 29554099 Free PMC article.
-
Biological Network Inference and analysis using SEBINI and CABIN.Methods Mol Biol. 2009;541:551-76. doi: 10.1007/978-1-59745-243-4_24. Methods Mol Biol. 2009. PMID: 19381531 Review.
-
Artificial intelligence techniques for colorectal cancer drug metabolism: ontology and complex network.Curr Drug Metab. 2010 May;11(4):347-68. doi: 10.2174/138920010791514289. Curr Drug Metab. 2010. PMID: 20446907 Review.
Cited by
-
Education, collaboration, and innovation: intelligent biology and medicine in the era of big data.BMC Genomics. 2015;16 Suppl 7(Suppl 7):S1. doi: 10.1186/1471-2164-16-S7-S1. Epub 2015 Jun 11. BMC Genomics. 2015. PMID: 26099197 Free PMC article.
-
Data integration to prioritize drugs using genomics and curated data.BioData Min. 2016 May 26;9:21. doi: 10.1186/s13040-016-0097-1. eCollection 2016. BioData Min. 2016. PMID: 27231484 Free PMC article.
-
Male mouse skeletal muscle lacking HuR shows enhanced glucose disposal at a young age.Front Physiol. 2025 Feb 19;15:1468369. doi: 10.3389/fphys.2024.1468369. eCollection 2024. Front Physiol. 2025. PMID: 40046510 Free PMC article.
-
Identification of Novel Genes Associated with Partial Resistance to Aphanomyces Root Rot in Field Pea by BSR-Seq Analysis.Int J Mol Sci. 2022 Aug 28;23(17):9744. doi: 10.3390/ijms23179744. Int J Mol Sci. 2022. PMID: 36077139 Free PMC article.
-
Identification of protein complexes associated with myocardial infarction using a bioinformatics approach.Mol Med Rep. 2018 Oct;18(4):3569-3576. doi: 10.3892/mmr.2018.9414. Epub 2018 Aug 21. Mol Med Rep. 2018. PMID: 30132549 Free PMC article.
References
-
- Nishimura D. BioCarta. Biotech Software & Internet Report. 2001;2(3):117–120. doi: 10.1089/152791601750294344. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

