Detection of selection signatures in Piemontese and Marchigiana cattle, two breeds with similar production aptitudes but different selection histories
- PMID: 26100250
- PMCID: PMC4476081
- DOI: 10.1186/s12711-015-0128-2
Detection of selection signatures in Piemontese and Marchigiana cattle, two breeds with similar production aptitudes but different selection histories
Abstract
Background: Domestication and selection are processes that alter the pattern of within- and between-population genetic variability. They can be investigated at the genomic level by tracing the so-called selection signatures. Recently, sequence polymorphisms at the genome-wide level have been investigated in a wide range of animals. A common approach to detect selection signatures is to compare breeds that have been selected for different breeding goals (i.e. dairy and beef cattle). However, genetic variations in different breeds with similar production aptitudes and similar phenotypes can be related to differences in their selection history.
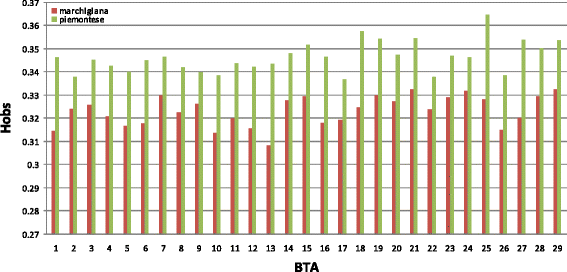
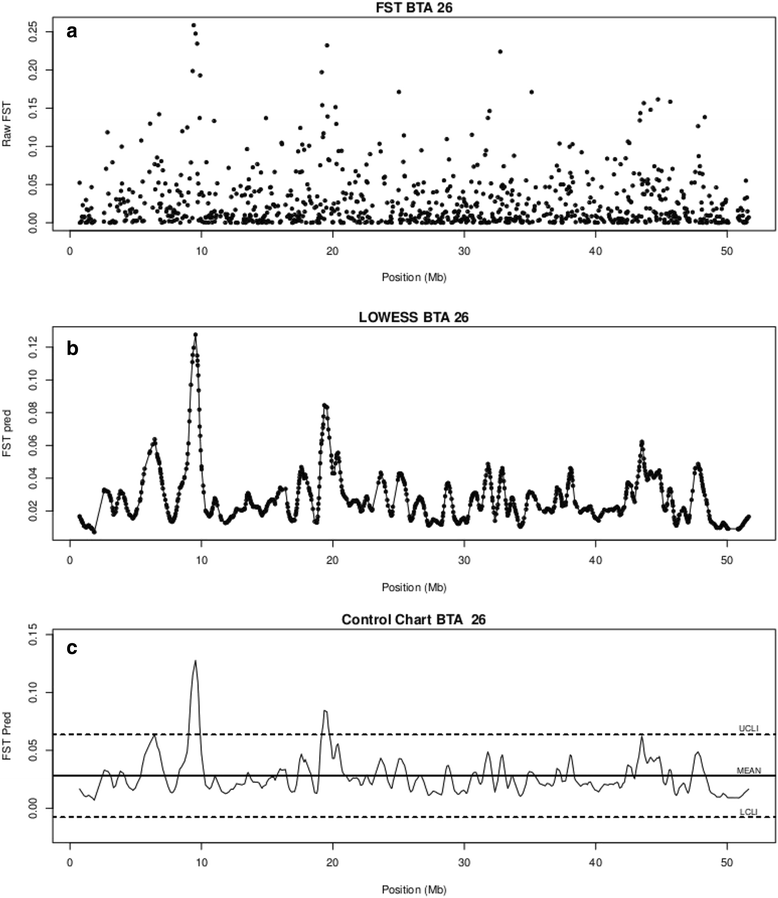
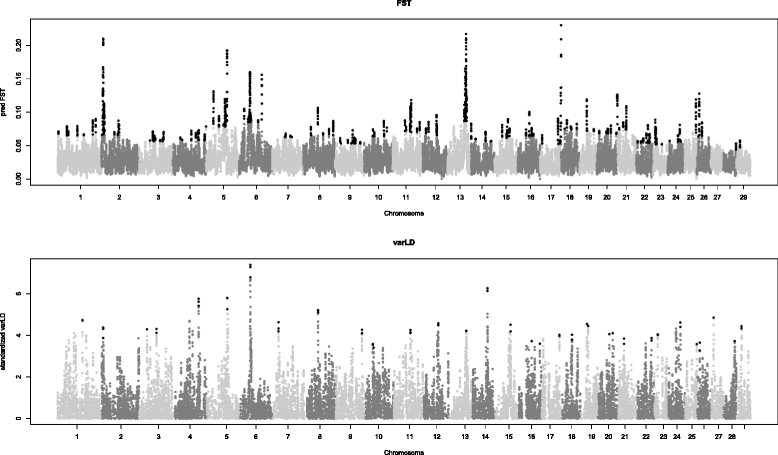
Methods: In this study, we investigated selection signatures between two Italian beef cattle breeds, Piemontese and Marchigiana, using genotyping data that was obtained with the Illumina BovineSNP50 BeadChip. The comparison was based on the fixation index (Fst), combined with a locally weighted scatterplot smoothing (LOWESS) regression and a control chart approach. In addition, analyses of Fst were carried out to confirm candidate genes. In particular, data were processed using the varLD method, which compares the regional variation of linkage disequilibrium between populations.
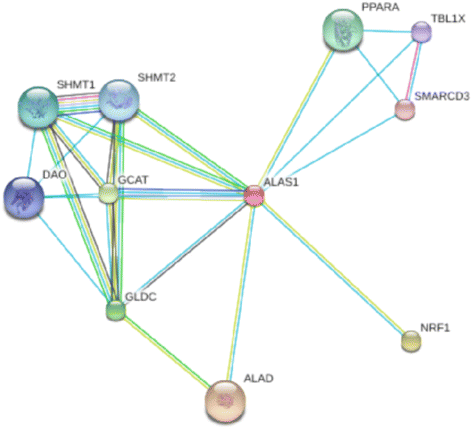
Results: Genome scans confirmed the presence of selective sweeps in the genomic regions that harbour candidate genes that are known to affect productive traits in cattle such as DGAT1, ABCG2, CAPN3, MSTN and FTO. In addition, several new putative candidate genes (for example ALAS1, ABCB8, ACADS and SOD1) were detected.
Conclusions: This study provided evidence on the different selection histories of two cattle breeds and the usefulness of genomic scans to detect selective sweeps even in cattle breeds that are bred for similar production aptitudes.
Figures
Similar articles
-
Use of canonical discriminant analysis to study signatures of selection in cattle.Genet Sel Evol. 2016 Aug 12;48(1):58. doi: 10.1186/s12711-016-0236-7. Genet Sel Evol. 2016. PMID: 27521154 Free PMC article.
-
Detection of selection signatures in dairy and beef cattle using high-density genomic information.Genet Sel Evol. 2015 Jun 19;47(1):49. doi: 10.1186/s12711-015-0127-3. Genet Sel Evol. 2015. PMID: 26089079 Free PMC article.
-
Use of locally weighted scatterplot smoothing (LOWESS) regression to study selection signatures in Piedmontese and Italian Brown cattle breeds.Anim Genet. 2014 Feb;45(1):1-11. doi: 10.1111/age.12076. Epub 2013 Jul 25. Anim Genet. 2014. PMID: 23889699
-
Assessing genomic diversity and signatures of selection in Jiaxian Red cattle using whole-genome sequencing data.BMC Genomics. 2021 Jan 9;22(1):43. doi: 10.1186/s12864-020-07340-0. BMC Genomics. 2021. PMID: 33421990 Free PMC article. Review.
-
Review: Opportunities and challenges for small populations of dairy cattle in the era of genomics.Animal. 2016 Jun;10(6):1050-60. doi: 10.1017/S1751731116000410. Epub 2016 Mar 9. Animal. 2016. PMID: 26957010 Review.
Cited by
-
A Meta-Assembly of Selection Signatures in Cattle.PLoS One. 2016 Apr 5;11(4):e0153013. doi: 10.1371/journal.pone.0153013. eCollection 2016. PLoS One. 2016. PMID: 27045296 Free PMC article.
-
Use of canonical discriminant analysis to study signatures of selection in cattle.Genet Sel Evol. 2016 Aug 12;48(1):58. doi: 10.1186/s12711-016-0236-7. Genet Sel Evol. 2016. PMID: 27521154 Free PMC article.
-
Association of DGAT1 With Cattle, Buffalo, Goat, and Sheep Milk and Meat Production Traits.Front Vet Sci. 2021 Aug 16;8:712470. doi: 10.3389/fvets.2021.712470. eCollection 2021. Front Vet Sci. 2021. PMID: 34485439 Free PMC article. Review.
-
Within- and between-Breed Selection Signatures in the Original and Improved Valachian Sheep.Animals (Basel). 2022 May 25;12(11):1346. doi: 10.3390/ani12111346. Animals (Basel). 2022. PMID: 35681809 Free PMC article.
-
Combining Landscape Genomics and Ecological Modelling to Investigate Local Adaptation of Indigenous Ugandan Cattle to East Coast Fever.Front Genet. 2018 Oct 3;9:385. doi: 10.3389/fgene.2018.00385. eCollection 2018. Front Genet. 2018. PMID: 30333851 Free PMC article.
References
-
- Felius M. Cattle breeds: an encyclopedia. 1. Misset: Doetinchem; 1995.
-
- Schwarzenbacher H, Dolezal M, Flisikowski K, Seefried F, Wurmser C, Schlötterer C, et al. Combining evidence of selection with association analysis increases power to detect regions influencing complex traits in dairy cattle. BMC Genomics. 2012;13:48. doi: 10.1186/1471-2164-13-48. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

