Linked genetic variants on chromosome 10 control ear morphology and body mass among dog breeds
- PMID: 26100605
- PMCID: PMC4477608
- DOI: 10.1186/s12864-015-1702-2
Linked genetic variants on chromosome 10 control ear morphology and body mass among dog breeds
Abstract
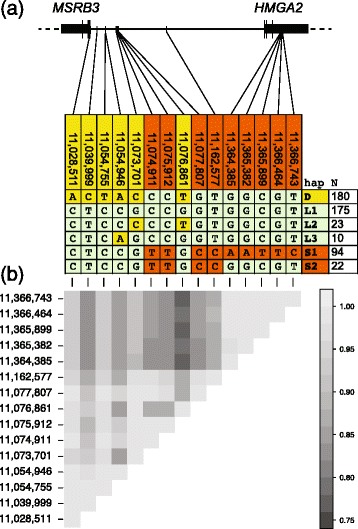
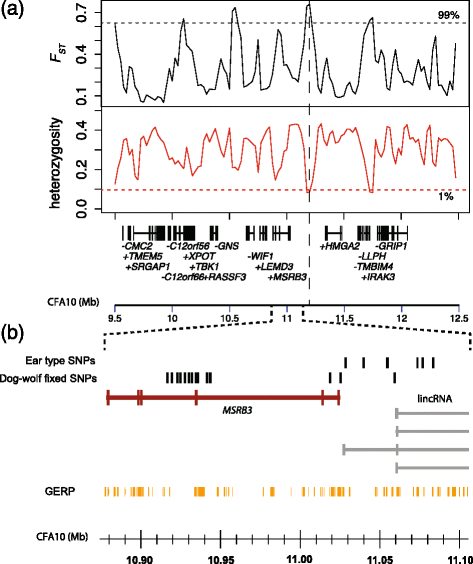
Background: The domestic dog is a rich resource for mapping the genetic components of phenotypic variation due to its unique population history involving strong artificial selection. Genome-wide association studies have revealed a number of chromosomal regions where genetic variation associates with morphological characters that typify dog breeds. A region on chromosome 10 is among those with the highest levels of genetic differentiation between dog breeds and is associated with body mass and ear morphology, a common motif of animal domestication. We characterised variation in this region to uncover haplotype structure and identify candidate functional variants.
Results: We first identified SNPs that strongly associate with body mass and ear type by comparing sequence variation in a 3 Mb region between 19 breeds with a variety of phenotypes. We next genotyped a subset of 123 candidate SNPs in 288 samples from 46 breeds to identify the variants most highly associated with phenotype and infer haplotype structure. A cluster of SNPs that associate strongly with the drop ear phenotype is located within a narrow interval downstream of the gene MSRB3, which is involved in human hearing. These SNPs are in strong genetic linkage with another set of variants that correlate with body mass within the gene HMGA2, which affects human height. In addition we find evidence that this region has been under selection during dog domestication, and identify a cluster of SNPs within MSRB3 that are highly differentiated between dogs and wolves.
Conclusions: We characterise genetically linked variants that potentially influence ear type and body mass in dog breeds, both key traits that have been modified by selective breeding that may also be important for domestication. The finding that variants on long haplotypes have effects on more than one trait suggests that genetic linkage can be an important determinant of the phenotypic response to selection in domestic animals.
Figures


Similar articles
-
Identification of genomic regions associated with phenotypic variation between dog breeds using selection mapping.PLoS Genet. 2011 Oct;7(10):e1002316. doi: 10.1371/journal.pgen.1002316. Epub 2011 Oct 13. PLoS Genet. 2011. PMID: 22022279 Free PMC article.
-
Ear type in sheep is associated with the MSRB3 locus.Anim Genet. 2020 Dec;51(6):968-972. doi: 10.1111/age.12994. Epub 2020 Aug 17. Anim Genet. 2020. PMID: 32805068
-
Identification of genomic variants putatively targeted by selection during dog domestication.BMC Evol Biol. 2016 Jan 12;16:10. doi: 10.1186/s12862-015-0579-7. BMC Evol Biol. 2016. PMID: 26754411 Free PMC article.
-
Toward understanding dog evolutionary and domestication history.C R Biol. 2011 Mar;334(3):190-6. doi: 10.1016/j.crvi.2010.12.011. Epub 2011 Feb 1. C R Biol. 2011. PMID: 21377613 Review.
-
Evolutionary genomics of dog domestication.Mamm Genome. 2012 Feb;23(1-2):3-18. doi: 10.1007/s00335-011-9386-7. Epub 2012 Jan 22. Mamm Genome. 2012. PMID: 22270221 Review.
Cited by
-
Genomic Uniqueness of Local Sheep Breeds From Morocco.Front Genet. 2021 Dec 2;12:723599. doi: 10.3389/fgene.2021.723599. eCollection 2021. Front Genet. 2021. PMID: 34925440 Free PMC article.
-
A scan for genes associated with cancer mortality and longevity in pedigree dog breeds.Mamm Genome. 2020 Aug;31(7-8):215-227. doi: 10.1007/s00335-020-09845-1. Epub 2020 Jul 13. Mamm Genome. 2020. PMID: 32661568 Free PMC article.
-
Ancestry-inclusive dog genomics challenges popular breed stereotypes.Science. 2022 Apr 29;376(6592):eabk0639. doi: 10.1126/science.abk0639. Epub 2022 Apr 29. Science. 2022. PMID: 35482869 Free PMC article.
-
Large scale across-breed genome-wide association study reveals a variant in HMGA2 associated with inguinal cryptorchidism risk in dogs.PLoS One. 2022 May 26;17(5):e0267604. doi: 10.1371/journal.pone.0267604. eCollection 2022. PLoS One. 2022. PMID: 35617214 Free PMC article.
-
Genome-Wide Association Studies Reveal Neurological Genes for Dog Herding, Predation, Temperament, and Trainability Traits.Front Vet Sci. 2021 Jul 21;8:693290. doi: 10.3389/fvets.2021.693290. eCollection 2021. Front Vet Sci. 2021. PMID: 34368281 Free PMC article.
References
-
- Pang J-F, Kluetsch C, Zou X-J, Zhang A, Luo L-Y, Angleby H, Ardalan A, Ekström C, Sköllermo A, Lundeberg J, Matsumura S, Leitner T, Zhang Y-P, Savolainen P. mtDNA data indicate a single origin for dogs south of Yangtze River, less than 16,300 years ago, from numerous wolves. Mol Biol Evol. 2009;26:2849–2864. doi: 10.1093/molbev/msp195. - DOI - PMC - PubMed
-
- Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, Kamal M, Clamp M, Chang JL, Kulbokas EJ, Zody MC, Mauceli E, Xie X, Breen M, Wayne RK, Ostrander EA, Ponting CP, Galibert F, Smith DR, DeJong PJ, Kirkness E, Alvarez P, Biagi T, Brockman W, Butler J, Chin CW, Cook A, Cuff J, Daly MJ, DeCaprio D, Gnerre S, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–19. doi: 10.1038/nature04338. - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

