Simulation-guided DNA probe design for consistently ultraspecific hybridization
- PMID: 26100802
- PMCID: PMC4479422
- DOI: 10.1038/nchem.2266
Simulation-guided DNA probe design for consistently ultraspecific hybridization
Abstract
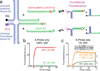
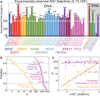
Hybridization of complementary sequences is one of the central tenets of nucleic acid chemistry; however, the unintended binding of closely related sequences limits the accuracy of hybridization-based approaches to analysing nucleic acids. Thermodynamics-guided probe design and empirical optimization of the reaction conditions have been used to enable the discrimination of single-nucleotide variants, but typically these approaches provide only an approximately 25-fold difference in binding affinity. Here we show that simulations of the binding kinetics are both necessary and sufficient to design nucleic acid probe systems with consistently high specificity as they enable the discovery of an optimal combination of thermodynamic parameters. Simulation-guided probe systems designed against 44 sequences of different target single-nucleotide variants showed between a 200- and 3,000-fold (median 890) higher binding affinity than their corresponding wild-type sequences. As a demonstration of the usefulness of this simulation-guided design approach, we developed probes that, in combination with PCR amplification, detect low concentrations of variant alleles (1%) in human genomic DNA.
Figures




Similar articles
-
An Engineered Kinetic Amplification Mechanism for Single Nucleotide Variant Discrimination by DNA Hybridization Probes.J Am Chem Soc. 2016 Apr 20;138(15):5076-86. doi: 10.1021/jacs.6b00277. Epub 2016 Apr 11. J Am Chem Soc. 2016. PMID: 27010123
-
Hybridization kinetics and thermodynamics of molecular beacons.Nucleic Acids Res. 2003 Feb 15;31(4):1319-30. doi: 10.1093/nar/gkg212. Nucleic Acids Res. 2003. PMID: 12582252 Free PMC article.
-
Simulation-Guided Rational Design of DNA Probe for Accurate Discrimination of Single-Nucleotide Variants Based on "Hill-Type" Cooperativity.Anal Chem. 2023 Feb 7;95(5):2893-2900. doi: 10.1021/acs.analchem.2c04446. Epub 2023 Jan 25. Anal Chem. 2023. PMID: 36695821
-
DNA probes: applications of the principles of nucleic acid hybridization.Crit Rev Biochem Mol Biol. 1991;26(3-4):227-59. doi: 10.3109/10409239109114069. Crit Rev Biochem Mol Biol. 1991. PMID: 1718662 Review.
-
Strategies for optimizing DNA hybridization on surfaces.Anal Biochem. 2014 Jan 1;444:41-6. doi: 10.1016/j.ab.2013.09.032. Epub 2013 Oct 9. Anal Biochem. 2014. PMID: 24121011 Review.
Cited by
-
A guide to accelerated direct digital counting of single nucleic acid molecules by FRET-based intramolecular kinetic fingerprinting.Methods. 2022 Jan;197:63-73. doi: 10.1016/j.ymeth.2021.06.014. Epub 2021 Jun 25. Methods. 2022. PMID: 34182140 Free PMC article. Review.
-
Cross-Inhibitor: a time-sensitive molecular circuit based on DNA strand displacement.Nucleic Acids Res. 2020 Nov 4;48(19):10691-10701. doi: 10.1093/nar/gkaa835. Nucleic Acids Res. 2020. PMID: 33045746 Free PMC article.
-
PAM-independent ultra-specific activation of CRISPR-Cas12a via sticky-end dsDNA.Nucleic Acids Res. 2022 Dec 9;50(22):12674-12688. doi: 10.1093/nar/gkac1144. Nucleic Acids Res. 2022. PMID: 36484104 Free PMC article.
-
Programmable autonomous synthesis of single-stranded DNA.Nat Chem. 2018 Feb;10(2):155-164. doi: 10.1038/nchem.2872. Epub 2017 Nov 6. Nat Chem. 2018. PMID: 29359755 Free PMC article.
-
Highly multiplexed rapid DNA detection with single-nucleotide specificity via convective PCR in a portable device.Nat Biomed Eng. 2021 Jul;5(7):702-712. doi: 10.1038/s41551-021-00755-4. Epub 2021 Jul 1. Nat Biomed Eng. 2021. PMID: 34211146
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

