Generating new prions by targeted mutation or segment duplication
- PMID: 26100899
- PMCID: PMC4507246
- DOI: 10.1073/pnas.1501072112
Generating new prions by targeted mutation or segment duplication
Abstract
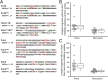
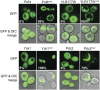
Yeasts contain various protein-based genetic elements, termed prions, that result from the structural conversion of proteins into self-propagating amyloid forms. Most yeast prion proteins contain glutamine/asparagine (Q/N)-rich prion domains that drive prion activity. Here, we explore two mechanisms by which new prion domains could evolve. First, it has been proposed that mutation and natural selection will tend to result in proteins with aggregation propensities just low enough to function under physiological conditions and thus that a small number of mutations are often sufficient to cause aggregation. We hypothesized that if the ability to form prion aggregates was a sufficiently generic feature of Q/N-rich domains, many nonprion Q/N-rich domains might similarly have aggregation propensities on the edge of prion formation. Indeed, we tested four yeast Q/N-rich domains that had no detectable aggregation activity; in each case, a small number of rationally designed mutations were sufficient to cause the proteins to aggregate and, for two of the domains, to create prion activity. Second, oligopeptide repeats are found in multiple prion proteins, and expansion of these repeats increases prion activity. However, it is unclear whether the effects of repeat expansion are unique to these specific sequences or are a generic result of adding additional aggregation-prone segments into a protein domain. We found that within nonprion Q/N-rich domains, repeating aggregation-prone segments in tandem was sufficient to create prion activity. Duplication of DNA elements is a common source of genetic variation and may provide a simple mechanism to rapidly evolve prion activity.
Keywords: Sup35; amyloid; prion; yeast.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
Controlling the prion propensity of glutamine/asparagine-rich proteins.Prion. 2015;9(5):347-54. doi: 10.1080/19336896.2015.1111506. Prion. 2015. PMID: 26555096 Free PMC article.
-
Emergence and evolution of yeast prion and prion-like proteins.BMC Evol Biol. 2016 Jan 25;16:24. doi: 10.1186/s12862-016-0594-3. BMC Evol Biol. 2016. PMID: 26809710 Free PMC article.
-
Effects of Q/N-rich, polyQ, and non-polyQ amyloids on the de novo formation of the [PSI+] prion in yeast and aggregation of Sup35 in vitro.Proc Natl Acad Sci U S A. 2004 Aug 31;101(35):12934-9. doi: 10.1073/pnas.0404968101. Epub 2004 Aug 23. Proc Natl Acad Sci U S A. 2004. PMID: 15326312 Free PMC article.
-
Prions of yeast as heritable amyloidoses.J Struct Biol. 2000 Jun;130(2-3):310-22. doi: 10.1006/jsbi.2000.4250. J Struct Biol. 2000. PMID: 10940235 Review.
-
The story of stolen chaperones: how overexpression of Q/N proteins cures yeast prions.Prion. 2013 Jul-Aug;7(4):294-300. doi: 10.4161/pri.26021. Epub 2013 Aug 7. Prion. 2013. PMID: 23924684 Free PMC article. Review.
Cited by
-
Amino acid composition predicts prion activity.PLoS Comput Biol. 2017 Apr 10;13(4):e1005465. doi: 10.1371/journal.pcbi.1005465. eCollection 2017 Apr. PLoS Comput Biol. 2017. PMID: 28394888 Free PMC article.
-
Controlling the prion propensity of glutamine/asparagine-rich proteins.Prion. 2015;9(5):347-54. doi: 10.1080/19336896.2015.1111506. Prion. 2015. PMID: 26555096 Free PMC article.
-
Effects of Mutations on the Aggregation Propensity of the Human Prion-Like Protein hnRNPA2B1.Mol Cell Biol. 2017 Mar 31;37(8):e00652-16. doi: 10.1128/MCB.00652-16. Print 2017 Apr 15. Mol Cell Biol. 2017. PMID: 28137911 Free PMC article.
-
Prion-like domains as epigenetic regulators, scaffolds for subcellular organization, and drivers of neurodegenerative disease.Brain Res. 2016 Sep 15;1647:9-18. doi: 10.1016/j.brainres.2016.02.037. Epub 2016 Mar 18. Brain Res. 2016. PMID: 26996412 Free PMC article. Review.
-
Anti-Prion Systems in Saccharomyces cerevisiae.J Neurochem. 2025 Mar;169(3):e70045. doi: 10.1111/jnc.70045. J Neurochem. 2025. PMID: 40130511 Free PMC article. Review.
References
-
- Sipe JD, Cohen AS. Review: History of the amyloid fibril. J Struct Biol. 2000;130(2–3):88–98. - PubMed
-
- Kisilevsky R, Fraser PE. A beta amyloidogenesis: Unique, or variation on a systemic theme? Crit Rev Biochem Mol Biol. 1997;32(5):361–404. - PubMed
-
- Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216(4542):136–144. - PubMed
-
- Fowler DM, Koulov AV, Balch WE, Kelly JW. Functional amyloid—from bacteria to humans. Trends Biochem Sci. 2007;32(5):217–224. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

