Structural insights into the recognition of cisplatin and AAF-dG lesion by Rad14 (XPA)
- PMID: 26100901
- PMCID: PMC4500287
- DOI: 10.1073/pnas.1508509112
Structural insights into the recognition of cisplatin and AAF-dG lesion by Rad14 (XPA)
Abstract
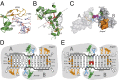
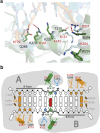
Nucleotide excision repair (NER) is responsible for the removal of a large variety of structurally diverse DNA lesions. Mutations of the involved proteins cause the xeroderma pigmentosum (XP) cancer predisposition syndrome. Although the general mechanism of the NER process is well studied, the function of the XPA protein, which is of central importance for successful NER, has remained enigmatic. It is known, that XPA binds kinked DNA structures and that it interacts also with DNA duplexes containing certain lesions, but the mechanism of interactions is unknown. Here we present two crystal structures of the DNA binding domain (DBD) of the yeast XPA homolog Rad14 bound to DNA with either a cisplatin lesion (1,2-GG) or an acetylaminofluorene adduct (AAF-dG). In the structures, we see that two Rad14 molecules bind to the duplex, which induces DNA melting of the duplex remote from the lesion. Each monomer interrogates the duplex with a β-hairpin, which creates a 13mer duplex recognition motif additionally characterized by a sharp 70° DNA kink at the position of the lesion. Although the 1,2-GG lesion stabilizes the kink due to the covalent fixation of the crosslinked dG bases at a 90° angle, the AAF-dG fully intercalates into the duplex to stabilize the kinked structure.
Keywords: AAF; NER; Rad14; XPA; cisplatin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




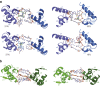
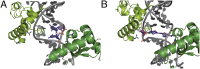

References
-
- Cleaver JE. Common pathways for ultraviolet skin carcinogenesis in the repair and replication defective groups of xeroderma pigmentosum. J Dermatol Sci. 2000;23(1):1–11. - PubMed
-
- Berneburg M, Lehmann AR. Xeroderma pigmentosum and related disorders: Defects in DNA repair and transcription. Adv Genet. 2001;43:71–102. - PubMed
-
- Naegeli H, Sugasawa K. The xeroderma pigmentosum pathway: Decision tree analysis of DNA quality. DNA Repair (Amst) 2011;10(7):673–683. - PubMed
-
- Sancar A. Mechanisms of DNA excision repair. Science. 1994;266(5193):1954–1956. - PubMed
-
- Geacintov NE, et al. Thermodynamic and structural factors in the removal of bulky DNA adducts by the nucleotide excision repair machinery. Biopolymers. 2002;65(3):202–210. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

