The evolution of Campylobacter jejuni and Campylobacter coli
- PMID: 26101080
- PMCID: PMC4526750
- DOI: 10.1101/cshperspect.a018119
The evolution of Campylobacter jejuni and Campylobacter coli
Abstract
The global significance of Campylobacter jejuni and Campylobacter coli as gastrointestinal human pathogens has motivated numerous studies to characterize their population biology and evolution. These bacteria are a common component of the intestinal microbiota of numerous bird and mammal species and cause disease in humans, typically via consumption of contaminated meat products, especially poultry meat. Sequence-based molecular typing methods, such as multilocus sequence typing (MLST) and whole genome sequencing (WGS), have been instructive for understanding the epidemiology and evolution of these bacteria and how phenotypic variation relates to the high degree of genetic structuring in C. coli and C. jejuni populations. Here, we describe aspects of the relatively short history of coevolution between humans and pathogenic Campylobacter, by reviewing research investigating how mutation and lateral or horizontal gene transfer (LGT or HGT, respectively) interact to create the observed population structure. These genetic changes occur in a complex fitness landscape with divergent ecologies, including multiple host species, which can lead to rapid adaptation, for example, through frame-shift mutations that alter gene expression or the acquisition of novel genetic elements by HGT. Recombination is a particularly strong evolutionary force in Campylobacter, leading to the emergence of new lineages and even large-scale genome-wide interspecies introgression between C. jejuni and C. coli. The increasing availability of large genome datasets is enhancing understanding of Campylobacter evolution through the application of methods, such as genome-wide association studies, but MLST-derived clonal complex designations remain a useful method for describing population structure.
Copyright © 2015 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures
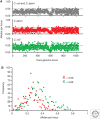
References
-
- Allos B. 2001. Campylobacter jejuni infections: Update on emerging issues and trends. Clin Infect Dis 32: 1201–1206. - PubMed
-
- Ammerman AJ, Cavalii-Sforza LL. 1984. The neolithic transition and the genetics of populations in Europe. Princeton University Press, Princeton, NJ.
-
- Bayliss CD, Bidmos FA, Anjum A, Manchev VT, Richards RL, Grossier JP, Wooldridge KG, Ketley JM, Barrow PA, Jones MA, et al. 2012. Phase variable genes of Campylobacter jejuni exhibit high mutation rates and specific mutational patterns but mutability is not the major determinant of population structure during host colonization. Nucleic Acids Res 40: 5876–5889. - PMC - PubMed
-
- Buckling A, Brockhurst MA, Travisano M, Rainey PB. 2007. Experimental adaptation to high and low quality environments under different scales of temporal variation. J Evol Biol 20: 296–300. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
