Pheromone-encoding mRNA is transported to the yeast mating projection by specific RNP granules
- PMID: 26101218
- PMCID: PMC4477862
- DOI: 10.1083/jcb.201408045
Pheromone-encoding mRNA is transported to the yeast mating projection by specific RNP granules
Abstract
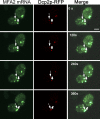
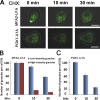
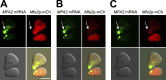
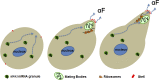
Association of messenger RNAs with large complexes such as processing bodies (PBs) plays a pivotal role in regulating their translation and decay. Little is known about other possible functions of these assemblies. Exposure of haploid yeast cells, carrying mating type "a," to "α pheromone" stimulates polarized growth resulting in a "shmoo" projection; it also induces synthesis of "a pheromone," encoded by MFA2. In this paper, we show that, in response to α pheromone, MFA2 mRNA is assembled with two types of granules; both contain some canonical PB proteins, yet they differ in size, localization, motility, and sensitivity to cycloheximide. Remarkably, one type is involved in mRNA transport to the tip of the shmoo, whereas the other-in local translation in the shmoo. Normal assembly of these granules is critical for their movement, localization, and for mating. Thus, MFA2 mRNAs are transported to the shmoo tip, in complex with PB-like particles, where they are locally translated.
© 2015 Aronov et al.
Figures


References
-
- Aronov S., Gelin-Licht R., Zipor G., Haim L., Safran E., and Gerst J.E.. 2007. mRNAs encoding polarity and exocytosis factors are cotransported with the cortical endoplasmic reticulum to the incipient bud in Saccharomyces cerevisiae. Mol. Cell. Biol. 27:3441–3455. 10.1128/MCB.01643-06 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

