Binding of STIL to Plk4 activates kinase activity to promote centriole assembly
- PMID: 26101219
- PMCID: PMC4477857
- DOI: 10.1083/jcb.201502088
Binding of STIL to Plk4 activates kinase activity to promote centriole assembly
Abstract
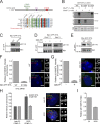
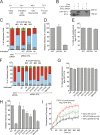
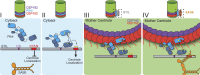
Centriole duplication occurs once per cell cycle in order to maintain control of centrosome number and ensure genome integrity. Polo-like kinase 4 (Plk4) is a master regulator of centriole biogenesis, but how its activity is regulated to control centriole assembly is unclear. Here we used gene editing in human cells to create a chemical genetic system in which endogenous Plk4 can be specifically inhibited using a cell-permeable ATP analogue. Using this system, we demonstrate that STIL localization to the centriole requires continued Plk4 activity. Most importantly, we show that direct binding of STIL activates Plk4 by promoting self-phosphorylation of the activation loop of the kinase. Plk4 subsequently phosphorylates STIL to promote centriole assembly in two steps. First, Plk4 activity promotes the recruitment of STIL to the centriole. Second, Plk4 primes the direct binding of STIL to the C terminus of SAS6. Our findings uncover a molecular basis for the timing of Plk4 activation through the cell cycle-regulated accumulation of STIL.
© 2015 Moyer et al.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

