Coordinated DNA Replication by the Bacteriophage T4 Replisome
- PMID: 26102578
- PMCID: PMC4488733
- DOI: 10.3390/v7062766
Coordinated DNA Replication by the Bacteriophage T4 Replisome
Abstract
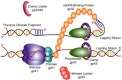
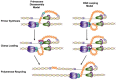
The T4 bacteriophage encodes eight proteins, which are sufficient to carry out coordinated leading and lagging strand DNA synthesis. These purified proteins have been used to reconstitute DNA synthesis in vitro and are a well-characterized model system. Recent work on the T4 replisome has yielded more detailed insight into the dynamics and coordination of proteins at the replication fork. Since the leading and lagging strands are synthesized in opposite directions, coordination of DNA synthesis as well as priming and unwinding is accomplished by several protein complexes. These protein complexes serve to link catalytic activities and physically tether proteins to the replication fork. Essential to both leading and lagging strand synthesis is the formation of a holoenzyme complex composed of the polymerase and a processivity clamp. The two holoenzymes form a dimer allowing the lagging strand polymerase to be retained within the replisome after completion of each Okazaki fragment. The helicase and primase also form a complex known as the primosome, which unwinds the duplex DNA while also synthesizing primers on the lagging strand. Future studies will likely focus on defining the orientations and architecture of protein complexes at the replication fork.
Keywords: DNA replication; T4 bacteriophage; replisome.
Figures
References
-
- Nossal N.G. Protein-protein interactions at a DNA replication fork: Bacteriophage T4 as a model. FASEB J. 1992;6:871–878. - PubMed
-
- Liu C., Burke R., Hibner U., Barry J., Alberts B. Cold Spring Harbor Symposia on Quantitative Biology. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY, USA: 1979. Probing DNA Replication Mechanisms with the T4 Bacteriophage in Vitro System; pp. 469–487. - PubMed
-
- Alberts B., Barry J., Bedinger P., Formosa T., Jongeneel C., Kreuzer K. Cold Spring Harbor Symposia on Quantitative Biology. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY, USA: 1983. Studies on DNA Replication in the Bacteriophage T4 in Vitro System; pp. 655–668. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

