MicroRNA396-Targeted SHORT VEGETATIVE PHASE Is Required to Repress Flowering and Is Related to the Development of Abnormal Flower Symptoms by the Phyllody Symptoms1 Effector
- PMID: 26103992
- PMCID: PMC4528741
- DOI: 10.1104/pp.15.00307
MicroRNA396-Targeted SHORT VEGETATIVE PHASE Is Required to Repress Flowering and Is Related to the Development of Abnormal Flower Symptoms by the Phyllody Symptoms1 Effector
Abstract
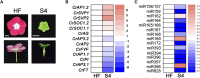
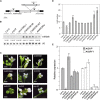
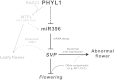
Leafy flowers are the major symptoms of peanut witches' broom (PnWB) phytoplasma infection in Catharanthus roseus. The orthologs of the phyllody symptoms1 (PHYL1) effector of PnWB from other species of phytoplasma can trigger the proteasomal degradation of several MADS box transcription factors, resulting in leafy flower formation. In contrast, the flowering negative regulator gene SHORT VEGETATIVE PHASE (SVP) was up-regulated in PnWB-infected C. roseus plants, but most microRNA (miRNA) genes had repressed expression. Coincidentally, transgenic Arabidopsis (Arabidopsis thaliana) plants expressing the PHYL1 gene of PnWB (PHYL1 plants), which show leafy flower phenotypes, up-regulate SVP of Arabidopsis (AtSVP) but repress a putative regulatory miRNA of AtSVP, miR396. However, the mechanism by which PHYL1 regulates AtSVP and miR396 is unknown, and the evidence of miR396-mediated AtSVP degradation is lacking. Here, we show that miR396 triggers AtSVP messenger RNA (mRNA) decay using genetic approaches, a reporter assay, and high-throughput degradome profiles. Genetic evidence indicates that PHYL1 plants and atmir396a-1 mutants have higher AtSVP accumulation, whereas the transgenic plants overexpressing MIR396 display lower AtSVP expression. The reporter assay indicated that target-site mutation results in decreasing the miR396-mediated repression efficiency. Moreover, degradome profiles revealed that miR396 triggers AtSVP mRNA decay rather than miRNA-mediated cleavage, implying that AtSVP caused miR396-mediated translation inhibition. We hypothesize that PHYL1 directly or indirectly interferes with miR396-mediated AtSVP mRNA decay and synergizes with other effects (e.g. MADS box transcription factor degradation), resulting in abnormal flower formation. We anticipate our findings to be a starting point for studying the posttranscriptional regulation of PHYL1 effectors in symptom development.
© 2015 American Society of Plant Biologists. All Rights Reserved.
Figures







References
-
- Bao M, Bian H, Zha Y, Li F, Sun Y, Bai B, Chen Z, Wang J, Zhu M, Han N (2014) miR396a-mediated basic helix-loop-helix transcription factor bHLH74 repression acts as a regulator for root growth in Arabidopsis seedlings. Plant Cell Physiol 55: 1343–1353 - PubMed
-
- Bowman JL, Alvarez J, Weigel D, Meyerowitz EM, Smyth DR (1993) Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development 119: 721–743
-
- Chang TC (2012) Identification of conserved microRNAs and their targets in PnWB phytoplasma induced leafy flower of Catharanthus roseus. PhD thesis. National Taiwan University, Taipei, Taiwan
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

