Mobile Bacterial Group II Introns at the Crux of Eukaryotic Evolution
- PMID: 26104554
- PMCID: PMC4394904
- DOI: 10.1128/microbiolspec.MDNA3-0050-2014
Mobile Bacterial Group II Introns at the Crux of Eukaryotic Evolution
Abstract
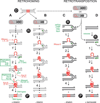
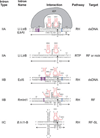
This review focuses on recent developments in our understanding of group II intron function, the relationships of these introns to retrotransposons and spliceosomes, and how their common features have informed thinking about bacterial group II introns as key elements in eukaryotic evolution. Reverse transcriptase-mediated and host factor-aided intron retrohoming pathways are considered along with retrotransposition mechanisms to novel sites in bacteria, where group II introns are thought to have originated. DNA target recognition and movement by target-primed reverse transcription infer an evolutionary relationship among group II introns, non-LTR retrotransposons, such as LINE elements, and telomerase. Additionally, group II introns are almost certainly the progenitors of spliceosomal introns. Their profound similarities include splicing chemistry extending to RNA catalysis, reaction stereochemistry, and the position of two divalent metals that perform catalysis at the RNA active site. There are also sequence and structural similarities between group II introns and the spliceosome's small nuclear RNAs (snRNAs) and between a highly conserved core spliceosomal protein Prp8 and a group II intron-like reverse transcriptase. It has been proposed that group II introns entered eukaryotes during bacterial endosymbiosis or bacterial-archaeal fusion, proliferated within the nuclear genome, necessitating evolution of the nuclear envelope, and fragmented giving rise to spliceosomal introns. Thus, these bacterial self-splicing mobile elements have fundamentally impacted the composition of extant eukaryotic genomes, including the human genome, most of which is derived from close relatives of mobile group II introns.
Figures




Similar articles
-
Bacterial group II introns generate genetic diversity by circularization and trans-splicing from a population of intron-invaded mRNAs.PLoS Genet. 2018 Nov 21;14(11):e1007792. doi: 10.1371/journal.pgen.1007792. eCollection 2018 Nov. PLoS Genet. 2018. PMID: 30462638 Free PMC article.
-
Organellar maturases: A window into the evolution of the spliceosome.Biochim Biophys Acta. 2015 Sep;1847(9):798-808. doi: 10.1016/j.bbabio.2015.01.009. Epub 2015 Jan 24. Biochim Biophys Acta. 2015. PMID: 25626174 Review.
-
Bacteria and Archaea Group II introns: additional mobile genetic elements in the environment.Environ Microbiol. 2003 Mar;5(3):143-51. doi: 10.1046/j.1462-2920.2003.00398.x. Environ Microbiol. 2003. PMID: 12588294 Review.
-
The mechanism of splicing as told by group II introns: Ancestors of the spliceosome.Biochim Biophys Acta Gene Regul Mech. 2019 Nov-Dec;1862(11-12):194390. doi: 10.1016/j.bbagrm.2019.06.001. Epub 2019 Jun 13. Biochim Biophys Acta Gene Regul Mech. 2019. PMID: 31202783 Review.
-
Mobile group II introns.Annu Rev Genet. 2004;38:1-35. doi: 10.1146/annurev.genet.38.072902.091600. Annu Rev Genet. 2004. PMID: 15568970 Review.
Cited by
-
Group II intron-like reverse transcriptases function in double-strand break repair.Cell. 2022 Sep 29;185(20):3671-3688.e23. doi: 10.1016/j.cell.2022.08.014. Epub 2022 Sep 15. Cell. 2022. PMID: 36113466 Free PMC article.
-
Revealing the high variability on nonconserved core and mobile elements of Austropuccinia psidii and other rust mitochondrial genomes.PLoS One. 2021 Mar 11;16(3):e0248054. doi: 10.1371/journal.pone.0248054. eCollection 2021. PLoS One. 2021. PMID: 33705433 Free PMC article.
-
The diversity of mtDNA rns introns among strains of Ophiostoma piliferum, Ophiostoma pluriannulatum and related species.Springerplus. 2016 Aug 24;5(1):1408. doi: 10.1186/s40064-016-3076-6. eCollection 2016. Springerplus. 2016. PMID: 27610327 Free PMC article.
-
Improved TGIRT-seq methods for comprehensive transcriptome profiling with decreased adapter dimer formation and bias correction.Sci Rep. 2019 May 28;9(1):7953. doi: 10.1038/s41598-019-44457-z. Sci Rep. 2019. PMID: 31138886 Free PMC article.
-
Molecular and Structural Characterization of an Immunopurified Telomerase from Leishmania major and the Effect of Telomerase Inhibitors.Microorganisms. 2025 Feb 7;13(2):357. doi: 10.3390/microorganisms13020357. Microorganisms. 2025. PMID: 40005724 Free PMC article.
References
-
- Lambowitz AM, Zimmerly S. Mobile group II introns. Annu Rev Genet. 2004;38:1–35. - PubMed
-
- Pyle AM, Lambowitz AM. The RNA World. 3rd ed. Cold Spring Harbor Laboratory Press; 2006. Group II introns: ribozymes that splice RNA and invade DNA; pp. 469–505.
-
- Toro N, Jiménez-Zurdo JI, García-Rodríguez FM. Bacterial group II introns: not just splicing. FEMS Microbiol Rev. 2007;31:342–358. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

