A genome-wide association study identifies multiple loci for variation in human ear morphology
- PMID: 26105758
- PMCID: PMC4491814
- DOI: 10.1038/ncomms8500
A genome-wide association study identifies multiple loci for variation in human ear morphology
Abstract
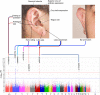
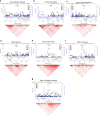
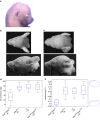
Here we report a genome-wide association study for non-pathological pinna morphology in over 5,000 Latin Americans. We find genome-wide significant association at seven genomic regions affecting: lobe size and attachment, folding of antihelix, helix rolling, ear protrusion and antitragus size (linear regression P values 2 × 10(-8) to 3 × 10(-14)). Four traits are associated with a functional variant in the Ectodysplasin A receptor (EDAR) gene, a key regulator of embryonic skin appendage development. We confirm expression of Edar in the developing mouse ear and that Edar-deficient mice have an abnormally shaped pinna. Two traits are associated with SNPs in a region overlapping the T-Box Protein 15 (TBX15) gene, a major determinant of mouse skeletal development. Strongest association in this region is observed for SNP rs17023457 located in an evolutionarily conserved binding site for the transcription factor Cartilage paired-class homeoprotein 1 (CART1), and we confirm that rs17023457 alters in vitro binding of CART1.
Figures

References
-
- Gleeson M. J. & Clarke R. C. Scott-Brown's Otorhinolaryngology: Head and Neck Surgery CRC Press (2008).
-
- Azaria R., Adler N., Silfen R., Regev D. & Hauben D. J. Morphometry of the adult human earlobe: a study of 547 subjects and clinical application. Plast. Reconstr. Surg. 111, 2398–2402 discussion 2403–2404 (2003). - PubMed
-
- Sforza C. et al. Age- and sex-related changes in the normal human ear. Forensic Sci. Int. 187, 110 e1–110 e7 (2009). - PubMed
-
- Alexander K. S., Stott D. J., Sivakumar B. & Kang N. A morphometric study of the human ear. J. Plast. Reconstr. Aesthet. Surg. 64, 41–47 (2011). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- RG/08/008/25291/BHF_/British Heart Foundation/United Kingdom
- BB/I021213/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MC_U142670371/MRC_/Medical Research Council/United Kingdom
- G0901388/MRC_/Medical Research Council/United Kingdom
- FS/13/6/29977/BHF_/British Heart Foundation/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

