S-layers at second glance? Altiarchaeal grappling hooks (hami) resemble archaeal S-layer proteins in structure and sequence
- PMID: 26106369
- PMCID: PMC4460559
- DOI: 10.3389/fmicb.2015.00543
S-layers at second glance? Altiarchaeal grappling hooks (hami) resemble archaeal S-layer proteins in structure and sequence
Abstract
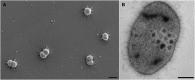
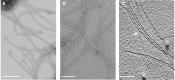
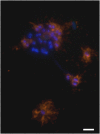
The uncultivated "Candidatus Altiarchaeum hamiconexum" (formerly known as SM1 Euryarchaeon) carries highly specialized nano-grappling hooks ("hami") on its cell surface. Until now little is known about the major protein forming these structured fibrous cell surface appendages, the genes involved or membrane anchoring of these filaments. These aspects were analyzed in depth in this study using environmental transcriptomics combined with imaging methods. Since a laboratory culture of this archaeon is not yet available, natural biofilm samples with high Ca. A. hamiconexum abundance were used for the entire analyses. The filamentous surface appendages spanned both membranes of the cell, which are composed of glycosyl-archaeol. The hami consisted of multiple copies of the same protein, the corresponding gene of which was identified via metagenome-mapped transcriptome analysis. The hamus subunit proteins, which are likely to self-assemble due to their predicted beta sheet topology, revealed no similiarity to known microbial flagella-, archaella-, fimbriae- or pili-proteins, but a high similarity to known S-layer proteins of the archaeal domain at their N-terminal region (44-47% identity). Our results provide new insights into the structure of the unique hami and their major protein and indicate their divergent evolution with S-layer proteins.
Keywords: S-layers; archaea; archaeal cell surface appendages; double-membrane; electron cryo-tomography; environmental transcriptomics; hami; nano-grappling hooks.
Figures


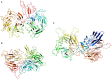
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

