Molecular mechanisms for the evolution of bacterial morphologies and growth modes
- PMID: 26106381
- PMCID: PMC4460556
- DOI: 10.3389/fmicb.2015.00580
Molecular mechanisms for the evolution of bacterial morphologies and growth modes
Abstract
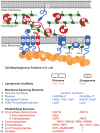
Bacteria exhibit a rich diversity of morphologies. Within this diversity, there is a uniformity of shape for each species that is replicated faithfully each generation, suggesting that bacterial shape is as selectable as any other biochemical adaptation. We describe the spatiotemporal mechanisms that target peptidoglycan synthesis to different subcellular zones to generate the rod-shape of model organisms Escherichia coli and Bacillus subtilis. We then demonstrate, using the related genera Caulobacter and Asticcacaulis as examples, how the modularity of the core components of the peptidoglycan synthesis machinery permits repositioning of the machinery to achieve different growth modes and morphologies. Finally, we highlight cases in which the mechanisms that underlie morphological evolution are beginning to be understood, and how they depend upon the expansion and diversification of the core components of the peptidoglycan synthesis machinery.
Keywords: Asticcacaulis; Caulobacter; FtsZ; MreB; bacterial morphology; bacterial shape; peptidoglycan synthesis.
Figures



References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

