Crystal structures of CRISPR-associated Csx3 reveal a manganese-dependent deadenylation exoribonuclease
- PMID: 26106927
- PMCID: PMC4615834
- DOI: 10.1080/15476286.2015.1051300
Crystal structures of CRISPR-associated Csx3 reveal a manganese-dependent deadenylation exoribonuclease
Abstract
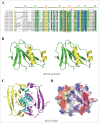
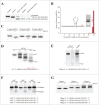
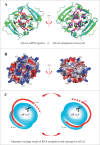
In prokaryotes, the CRISPR/Cas system is known to target and degrade invading phages and foreign genetic elements upon subsequent infection. However, the structure and function of many Cas proteins remain largely unknown, due to the high diversity of Cas proteins. Here we report 3 crystal structures of Archaeoglobus fulgidus Csx3 (AfCsx3) in free form, in complex with manganese ions and in complex with a single-stranded RNA (ssRNA) fragment, respectively. AfCsx3 harbors a ferredoxin-like fold and forms dimer both in the crystal and in solution. Our structure-based biochemical analysis demonstrates that the RNA binding sites and cleavage sites are located at 2 separate surfaces within the AfCsx3 dimer, suggesting a model to bind, tether and cleave the incoming RNA substrate. In addition, AfCsx3 displays robust 3'-deadenylase activity in the presence of manganese ions, which strongly suggests that AfCsx3 functions as a deadenylation exonuclease. Taken together, our results indicate that AfCsx3 is a Cas protein involved in RNA deadenylation and provide a framework for understanding the role of AfCsx3 in the Type III-B CRISPR/Cas system.
Keywords: Archaeoglobus fulgidus Csx3; CRISPR/Cas; deadenylation exonuclease; ferredoxin-like fold; manganese-dependent.
Figures


Comment in
-
Recognition of a pseudo-symmetric RNA tetranucleotide by Csx3, a new member of the CRISPR associated Rossmann fold superfamily.RNA Biol. 2016;13(2):254-7. doi: 10.1080/15476286.2015.1130209. RNA Biol. 2016. PMID: 26727591 Free PMC article.
References
-
- Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature 2012; 482:331-8; PMID:22337052; http://dx.doi.org/10.1038/nature10886 - DOI - PubMed
-
- Terns MP, Terns RM. CRISPR-based adaptive immune systems. Curr Opin Microbiol 2011; 14:321-7; PMID:21531607; http://dx.doi.org/10.1016/j.mib.2011.03.005 - DOI - PMC - PubMed
-
- Ruud Jansen, Jan DA. van Embden, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol 2002; 43:1565-75; PMID:11952905; http://dx.doi.org/10.1046/j.1365-2958.2002.02839.x - DOI - PubMed
-
- Shah SA, Hansen NR, Garrett RA. Distribution of CRISPR spacer matches in viruses and plasmids of crenarchaeal acidothermophiles and implications for their inhibitory mechanism. Biochem Soc Trans 2009; 37:23-8; PMID:19143596; http://dx.doi.org/10.1042/BST0370023 - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
