Dual fatty acid synthase and HER2 signaling blockade shows marked antitumor activity against breast cancer models resistant to anti-HER2 drugs
- PMID: 26107737
- PMCID: PMC4479882
- DOI: 10.1371/journal.pone.0131241
Dual fatty acid synthase and HER2 signaling blockade shows marked antitumor activity against breast cancer models resistant to anti-HER2 drugs
Abstract
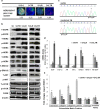
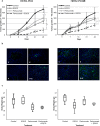
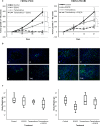
Blocking the enzyme Fatty Acid Synthase (FASN) leads to apoptosis of HER2-positive breast carcinoma cells. The hypothesis is that blocking FASN, in combination with anti-HER2 signaling agents, would be an effective antitumor strategy in preclinical HER2+ breast cancer models of trastuzumab and lapatinib resistance. We developed and molecularly characterized in vitro HER2+ models of resistance to trastuzumab (SKTR), lapatinib (SKLR) and both (SKLTR). The cellular interactions of combining anti-FASN polyphenolic compounds (EGCG and the synthetic G28UCM) with anti-HER2 signaling drugs (trastuzumab plus pertuzumab and temsirolimus) were analyzed. Tumor growth inhibition after treatment with EGCG, pertuzumab, temsirolimus or the combination was evaluated in two in vivo orthoxenopatients: one derived from a HER2+ patient and another from a patient who relapsed on trastuzumab and lapatinib-based therapy. SKTR, SKLR and SKLTR showed hyperactivation of EGFR and p-ERK1/2 and PI3KCA mutations. Dual-resistant cells (SKLTR) also showed hyperactivation of HER4 and recovered levels of p-AKT compared with mono-resistant cells. mTOR, p-mTOR and FASN expression remained stable in SKTR, SKLR and SKLTR. In vitro, anti-FASN compounds plus pertuzumab showed synergistic interactions in lapatinib- and dual- resistant cells and improved the results of pertuzumab plus trastuzumab co-treatment. FASN inhibitors combined with temsirolimus displayed the strongest synergistic interactions in resistant cells. In vivo, both orthoxenopatients showed strong response to the antitumor activity of the combination of EGCG with pertuzumab or temsirolimus, without signs of toxicity. We showed that the simultaneous blockade of FASN and HER2 pathways is effective in cells and in breast cancer models refractory to anti-HER2 therapies.
Conflict of interest statement
Figures


References
-
- Ferguson KM, Berger MB, Mendrola JM, Cho HS, Leahy DJ, Lemmon MA. EGF activates its receptor by removing interactions that autoinhibit ectodomain dimerization. Mol Cell. 2003;11(2):507–17. . - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

