Prevalence of tetracycline resistance genes among multi-drug resistant bacteria from selected water distribution systems in southwestern Nigeria
- PMID: 26108344
- PMCID: PMC4481114
- DOI: 10.1186/s12941-015-0093-1
Prevalence of tetracycline resistance genes among multi-drug resistant bacteria from selected water distribution systems in southwestern Nigeria
Erratum in
-
Erratum to: Prevalence of tetracycline resistance genes among multi-drug resistant bacteria from selected water distribution systems in southwestern Nigeria.Ann Clin Microbiol Antimicrob. 2015 Sep 11;14:41. doi: 10.1186/s12941-015-0099-8. Ann Clin Microbiol Antimicrob. 2015. PMID: 26362933 Free PMC article. No abstract available.
Abstract
Background: Antibiotic resistance genes [ARGs] in aquatic systems have drawn increasing attention they could be transferred horizontally to pathogenic bacteria. Water treatment plants (WTPs) are intended to provide quality and widely available water to the local populace they serve. However, WTPs in developing countries may not be dependable for clean water and they could serve as points of dissemination for antibiotic resistant bacteria. Only a few studies have investigated the occurrence of ARGs among these bacteria including tetracycline resistance genes in water distribution systems in Nigeria.
Methodology: Multi-drug resistant (MDR) bacteria, including resistance to tetracycline, were isolated from treated and untreated water distribution systems in southwest Nigeria. MDR bacteria were resistant to >3 classes of antibiotics based on break-point assays. Isolates were characterized using partial 16S rDNA sequencing and PCR assays for six tetracycline-resistance genes. Plasmid conjugation was evaluated using E. coli strain DH5α as the recipient strain.
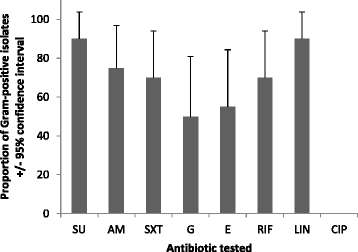
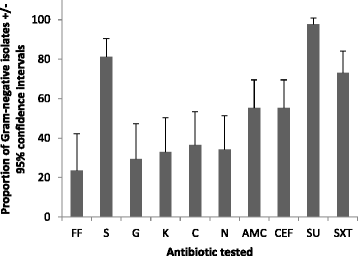
Results: Out of the 105 bacteria, 85 (81 %) and 20 (19 %) were Gram- negative or Gram- positive, respectively. Twenty-nine isolates carried at least one of the targeted tetracycline resistance genes including strains of Aeromonas, Alcaligenes, Bacillus, Klebsiella, Leucobacter, Morganella, Proteus and a sequence matching a previously uncultured bacteria. Tet(A) was the most prevalent (16/29) followed by tet(E) (4/29) and tet30 (2/29). Tet(O) was not detected in any of the isolates. Tet(A) was mostly found with Alcaligenes strains (9/10) and a combination of more than one resistance gene was observed only amongst Alcaligenes strains [tet(A) + tet30 (2/10), tet(A) + tet(E) (3/10), tet(E) + tet(M) (1/10), tet(E) + tet30 (1/10)]. Tet(A) was transferred by conjugation for five Alcaligenes and two E. coli isolates.
Conclusions: This study found a high prevalence of plasmid-encoded tet(A) among Alcaligenes isolates, raising the possibility that this strain could shuttle resistance plasmids to pathogenic bacteria.
Figures
References
-
- Bryskier A. Tetracyclines. In: Bryskier A, editor. Antimicrobial agents: antibacterials and antifungals. Washington: ASM Press; 2005. pp. 642–651.
-
- Stockstad ELR, Jukes TH, Pierce J, Page AC, Franklin AL. The multiple nature of animal protein factor. J Biol Chem. 1949;180:647–654. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

