The sorting protein PACS-2 promotes ErbB signalling by regulating recycling of the metalloproteinase ADAM17
- PMID: 26108729
- PMCID: PMC4481878
- DOI: 10.1038/ncomms8518
The sorting protein PACS-2 promotes ErbB signalling by regulating recycling of the metalloproteinase ADAM17
Abstract
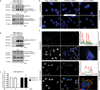
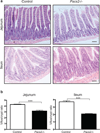
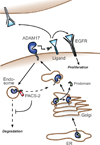
The metalloproteinase ADAM17 activates ErbB signalling by releasing ligands from the cell surface, a key step underlying epithelial development, growth and tumour progression. However, mechanisms acutely controlling ADAM17 cell-surface availability to modulate the extent of ErbB ligand release are poorly understood. Here, through a functional genome-wide siRNA screen, we identify the sorting protein PACS-2 as a regulator of ADAM17 trafficking and ErbB signalling. PACS-2 loss reduces ADAM17 cell-surface levels and ADAM17-dependent ErbB ligand shedding, without apparent effects on related proteases. PACS-2 co-localizes with ADAM17 on early endosomes and PACS-2 knockdown decreases the recycling and stability of internalized ADAM17. Hence, PACS-2 sustains ADAM17 cell-surface activity by diverting ADAM17 away from degradative pathways. Interestingly, Pacs2-deficient mice display significantly reduced levels of phosphorylated EGFR and intestinal proliferation. We suggest that this mechanism controlling ADAM17 cell-surface availability and EGFR signalling may play a role in intestinal homeostasis, with potential implications for cancer biology.
Figures





References
-
- Arribas J, Esselens C. ADAM17 as a therapeutic target in multiple diseases. Curr. Pharm. Des. 2009;15:2319–2335. - PubMed
-
- Blobel CP. ADAMs: key components in EGFR signalling and development. Nat. Rev. Mol. Cell Biol. 2005;6:32–43. - PubMed
-
- Murphy G. The ADAMs: signalling scissors in the tumour microenvironment. Nat. Rev. Cancer. 2008;8:929–941. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

