Tc-MYBPA an Arabidopsis TT2-like transcription factor and functions in the regulation of proanthocyanidin synthesis in Theobroma cacao
- PMID: 26109181
- PMCID: PMC4481123
- DOI: 10.1186/s12870-015-0529-y
Tc-MYBPA an Arabidopsis TT2-like transcription factor and functions in the regulation of proanthocyanidin synthesis in Theobroma cacao
Abstract
Background: The flavan-3-ols catechin and epicatechin, and their polymerized oligomers, the proanthocyanidins (PAs, also called condensed tannins), accumulate to levels of up to 15 % of the total weight of dry seeds of Theobroma cacao L. These compounds have been associated with several health benefits in humans. They also play important roles in pest and disease defense throughout the plant. In Arabidopsis, the R2R3 type MYB transcription factor TT2 regulates the major genes leading to the synthesis of PA.
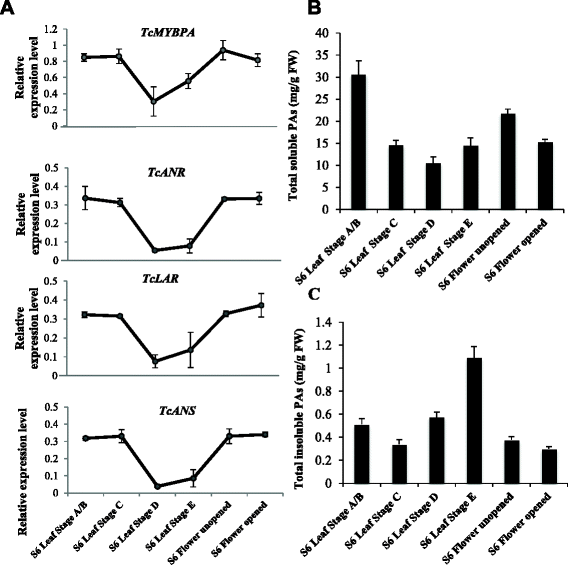
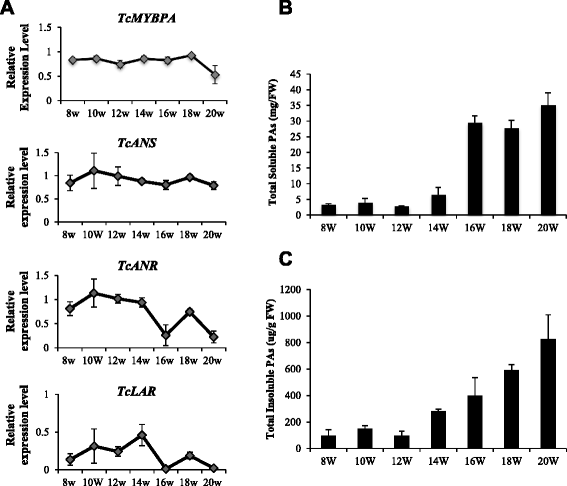
Results: To explore the transcriptional regulation of the PA synthesis pathway in cacao, we isolated and characterized an R2R3 type MYB transcription factor MYBPA from cacao. We examined the spatial and temporal gene expression patterns of the Tc-MYBPA gene and found it to be developmentally expressed in a manner consistent with its involvement in PAs and anthocyanin synthesis. Functional complementation of an Arabidopsis tt2 mutant with Tc-MYBPA suggested that it can functionally substitute the Arabidopsis TT2 gene. Interestingly, in addition to PA accumulation in seeds of the Tc-MYBPA expressing plants, we also observed an obvious increase of anthocyanidin accumulation in hypocotyls. We observed that overexpression of the Tc-MYBPA gene resulted in increased expression of several key genes encoding the major structural enzymes of the PA and anthocyanidin pathway, including DFR (dihydroflavanol reductase), LDOX (leucoanthocyanidin dioxygenase) and BAN (ANR, anthocyanidin reductase).
Conclusion: We conclude that the Tc-MYBPA gene that encodes an R2R3 type MYB transcription factor is an Arabidopsis TT2 like transcription factor, and may be involved in the regulation of both anthocyanin and PA synthesis in cacao. This research may provide molecular tools for breeding of cacao varieties with improved disease resistance and enhanced flavonoid profiles for nutritional and pharmaceutical applications.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

