Genome-wide mutation profiles of colorectal tumors and associated liver metastases at the exome and transcriptome levels
- PMID: 26109429
- PMCID: PMC4673155
- DOI: 10.18632/oncotarget.4246
Genome-wide mutation profiles of colorectal tumors and associated liver metastases at the exome and transcriptome levels
Abstract
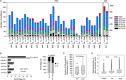
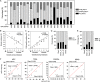
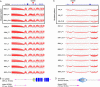
To characterize the mutation profiles of colorectal cancer (CRC) primary tumors (PTs) and liver metastases (CLMs), we performed both whole-exome and RNA sequencing. Ten significantly mutated genes, including BMI1, CARD11, and NRG1, were found in 34 CRCs with CLMs. We defined three mutation classes (Class 1 to 3) based on the absence or presence of mutations during liver metastasis. Most mutations were classified into Class 1 (shared between PTs and CLMs), suggesting the common clonal origin of PTs and CLMs. Class 1 was more strongly associated with the clinical characteristics of advanced cancer and was more frequently superimposed with chromosomal deletions in CLMs than Class 2 (PT-specific). The integration of exome and RNA sequencing revealed that variant-allele frequencies (VAFs) of mutations in the transcriptome tended to have stronger functional implications than those in the exome. For instance, VAFs of the TP53 and APC mutations in the transcriptome significantly correlated with the expression level of their target genes. Additionally, mutations with high functional impact were enriched with high VAFs in the CLM transcriptomes. We identified 11 mutation-associated splicing events in the CRC transcriptomes. Thus, the integration of the exome and the transcriptome may elucidate the underlying molecular events responsible for CLMs.
Keywords: RNA sequencing; colorectal cancer; exome sequencing; liver metastasis; somatic mutation.
Conflict of interest statement
No potential conflicts of interest were disclosed by all authors.
Figures




References
-
- Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–170. - PubMed
-
- Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM, Bos JL. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525–532. - PubMed
-
- Gylfe AE, Kondelin J, Turunen M, Ristolainen H, Katainen R, Pitkanen E, Kaasinen E, Rantanen V, Tanskanen T, Varjosalo M, Lehtonen H, Palin K, Taipale M, et al. Identification of candidate oncogenes in human colorectal cancers with microsatellite instability. Gastroenterology. 2013;145:540–543. e522. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- SRA/SRP041725
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

