Proteomic analysis of colon and rectal carcinoma using standard and customized databases
- PMID: 26110064
- PMCID: PMC4477697
- DOI: 10.1038/sdata.2015.22
Proteomic analysis of colon and rectal carcinoma using standard and customized databases
Erratum in
-
Corrigendum: Proteomic analysis of colon and rectal carcinoma using standard and customized databases.Sci Data. 2015 Jul 21;2:150037. doi: 10.1038/sdata.2015.37. eCollection 2015. Sci Data. 2015. PMID: 26217491 Free PMC article. No abstract available.
Abstract
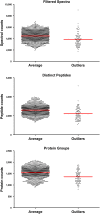
Understanding proteomic differences underlying the different phenotypic classes of colon and rectal carcinoma is important and may eventually lead to a better assessment of clinical behavior of these cancers. We here present a comprehensive description of the proteomic data obtained from 90 colon and rectal carcinomas previously subjected to genomic analysis by The Cancer Genome Atlas (TCGA). Here, the primary instrument files and derived secondary data files are compiled and presented in forms that will allow further analyses of the biology of colon and rectal carcinoma. We also discuss new challenges in processing these large proteomic datasets for relevant proteins and protein variants.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
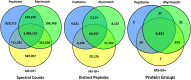
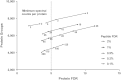
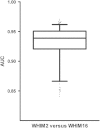
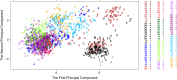

References
Data Citations
-
- Edwards N., Liebler D. C. 2014. ProteomeXchange . PXD001006
-
- 2014. Broad Institute Firehose TCGA sequence data for colon carcinoma . http://gdac.broadinstitute.org/runs/stddata__2013_05_23/data/COAD/201305...
-
- 2014. Broad Institute Firehose TCGA sequence data for rectal carcinoma . http://gdac.broadinstitute.org/runs/stddata__2013_05_23/data/READ/201305...
-
- Slebos R. J. C., Edwards N. 2015. ProteomeXchange . PXD002041
-
- Slebos R. J. C., Edwards N. 2015. ProteomeXchange . PXD002042
References
-
- Nesvizhskii A. I., Vitek O. & Aebersold R. Analysis and validation of proteomic data generated by tandem mass spectrometry. Nat. Methods 4, 787–797 (2007). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

