Alternative Okazaki Fragment Ligation Pathway by DNA Ligase III
- PMID: 26110316
- PMCID: PMC4488670
- DOI: 10.3390/genes6020385
Alternative Okazaki Fragment Ligation Pathway by DNA Ligase III
Abstract
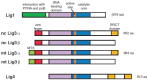
Higher eukaryotes have three types of DNA ligases: DNA ligase 1 (Lig1), DNA ligase 3 (Lig3) and DNA ligase 4 (Lig4). While Lig1 and Lig4 are present in all eukaryotes from yeast to human, Lig3 appears sporadically in evolution and is uniformly present only in vertebrates. In the classical, textbook view, Lig1 catalyzes Okazaki-fragment ligation at the DNA replication fork and the ligation steps of long-patch base-excision repair (BER), homologous recombination repair (HRR) and nucleotide excision repair (NER). Lig4 is responsible for DNA ligation at DNA double strand breaks (DSBs) by the classical, DNA-PKcs-dependent pathway of non-homologous end joining (C-NHEJ). Lig3 is implicated in a short-patch base excision repair (BER) pathway, in single strand break repair in the nucleus, and in all ligation requirements of the DNA metabolism in mitochondria. In this scenario, Lig1 and Lig4 feature as the major DNA ligases serving the most essential ligation needs of the cell, while Lig3 serves in the cell nucleus only minor repair roles. Notably, recent systematic studies in the chicken B cell line, DT40, involving constitutive and conditional knockouts of all three DNA ligases individually, as well as of combinations thereof, demonstrate that the current view must be revised. Results demonstrate that Lig1 deficient cells proliferate efficiently. Even Lig1/Lig4 double knockout cells show long-term viability and proliferate actively, demonstrating that, at least in DT40, Lig3 can perform all ligation reactions of the cellular DNA metabolism as sole DNA ligase. Indeed, in the absence of Lig1, Lig3 can efficiently support semi-conservative DNA replication via an alternative Okazaki-fragment ligation pathway. In addition, Lig3 can back up NHEJ in the absence of Lig4, and can support NER and HRR in the absence of Lig1. Supporting observations are available in less elaborate genetic models in mouse cells. Collectively, these observations raise Lig3 from a niche-ligase to a universal DNA ligase, which can potentially substitute or backup the repair and replication functions of all other DNA ligases in the cell nucleus. Thus, the old model of functionally dedicated DNA ligases is now replaced by one in which only Lig4 remains dedicated to C-NHEJ, with Lig1 and Lig3 showing an astounding functional flexibility and interchangeability for practically all nuclear ligation functions. The underlying mechanisms of Lig3 versus Lig1 utilization in DNA repair and replication are expected to be partly different and remain to be elucidated.
Keywords: DNA ligase 1; DNA ligase 3; DNA repair; DNA replication; DT40; Lig1; Lig3; Okazaki fragment.
Figures

References
-
- Husain I., Tomkinson A.E., Burkhart W.A., Moyer M.B., Ramos W., Mackey Z.B., Besterman J.M., Chen J. Purification and characterization of DNA ligase III from bovine testes. Homology with DNA ligase II and vaccinia DNA ligase. J. Biol. Chem. 1995;270:9683–9690. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

