Orientation of DNA Minicircles Balances Density and Topological Complexity in Kinetoplast DNA
- PMID: 26110537
- PMCID: PMC4482025
- DOI: 10.1371/journal.pone.0130998
Orientation of DNA Minicircles Balances Density and Topological Complexity in Kinetoplast DNA
Abstract
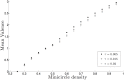
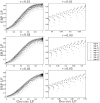
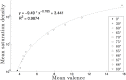
Kinetoplast DNA (kDNA), a unique mitochondrial structure common to trypanosomatid parasites, contains thousands of DNA minicircles that are densely packed and can be topologically linked into a chain mail-like network. Experimental data indicate that every minicircle in the network is, on average, singly linked to three other minicircles (i.e., has mean valence 3) before replication and to six minicircles in the late stages of replication. The biophysical factors that determine the topology of the network and its changes during the cell cycle remain unknown. Using a mathematical modeling approach, we previously showed that volume confinement alone can drive the formation of the network and that it induces a linear relationship between mean valence and minicircle density. Our modeling also predicted a minicircle valence two orders of magnitude greater than that observed in kDNA. To determine the factors that contribute to this discrepancy we systematically analyzed the relationship between the topological properties of the network (i.e., minicircle density and mean valence) and its biophysical properties such as DNA bending, electrostatic repulsion, and minicircle relative position and orientation. Significantly, our results showed that most of the discrepancy between the theoretical and experimental observations can be accounted for by the orientation of the minicircles with volume exclusion due to electrostatic interactions and DNA bending playing smaller roles. Our results are in agreement with the three dimensional kDNA organization model, initially proposed by Delain and Riou, in which minicircles are oriented almost perpendicular to the horizontal plane of the kDNA disk. We suggest that while minicircle confinement drives the formation of kDNA networks, it is minicircle orientation that regulates the topological complexity of the network.
Conflict of interest statement
Figures

Similar articles
-
The rotational dynamics of kinetoplast DNA replication.Mol Microbiol. 2007 May;64(3):676-90. doi: 10.1111/j.1365-2958.2007.05686.x. Mol Microbiol. 2007. PMID: 17462016
-
The attachment of minicircles to kinetoplast DNA networks during replication.Cell. 1993 Aug 27;74(4):703-11. doi: 10.1016/0092-8674(93)90517-t. Cell. 1993. PMID: 8395351
-
The topology of the kinetoplast DNA network.Cell. 1995 Jan 13;80(1):61-9. doi: 10.1016/0092-8674(95)90451-4. Cell. 1995. PMID: 7813018
-
The structure and replication of kinetoplast DNA.Annu Rev Microbiol. 1995;49:117-43. doi: 10.1146/annurev.mi.49.100195.001001. Annu Rev Microbiol. 1995. PMID: 8561456 Review.
-
The structure and replication of kinetoplast DNA.Curr Mol Med. 2004 Sep;4(6):623-47. doi: 10.2174/1566524043360096. Curr Mol Med. 2004. PMID: 15357213 Review.
Cited by
-
Human bloodsucking parasite in service of materials science.Proc Natl Acad Sci U S A. 2020 Jan 7;117(1):18-20. doi: 10.1073/pnas.1920496117. Epub 2019 Dec 26. Proc Natl Acad Sci U S A. 2020. PMID: 31879357 Free PMC article. No abstract available.
-
Unraveling the Influence of Topology and Spatial Confinement on Equilibrium and Relaxation Properties of Interlocked Ring Polymers.Macromolecules. 2024 Mar 21;57(7):3223-3233. doi: 10.1021/acs.macromol.3c02203. eCollection 2024 Apr 9. Macromolecules. 2024. PMID: 38616813 Free PMC article.
-
Direct monitoring of the stepwise condensation of kinetoplast DNA networks.Sci Rep. 2021 Jan 15;11(1):1501. doi: 10.1038/s41598-021-81045-6. Sci Rep. 2021. PMID: 33452335 Free PMC article.
-
Characterization of the novel mitochondrial genome segregation factor TAP110 in Trypanosoma brucei.J Cell Sci. 2021 Mar 8;134(5):jcs254300. doi: 10.1242/jcs.254300. J Cell Sci. 2021. PMID: 33589495 Free PMC article.
References
-
- World Health Organization. Research Priorities for Chagas Disease, Human African Trypanosomiasis and Leishmaniasis. World Health Organ Tech Rep Ser. 2012; 975: v–xii. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

