Bioinformatics analysis of thousands of TCGA tumors to determine the involvement of epigenetic regulators in human cancer
- PMID: 26110843
- PMCID: PMC4480953
- DOI: 10.1186/1471-2164-16-S8-S5
Bioinformatics analysis of thousands of TCGA tumors to determine the involvement of epigenetic regulators in human cancer
Abstract
Background: Many cancer cells show distorted epigenetic landscapes. The Cancer Genome Atlas (TCGA) project profiles thousands of tumors, allowing the discovery of somatic alterations in the epigenetic machinery and the identification of potential cancer drivers among members of epigenetic protein families.
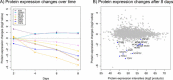
Methods: We integrated mutation, expression, and copy number data from 5943 tumors from 13 cancer types to train a classification model that predicts the likelihood of being an oncogene (OG), tumor suppressor (TSG) or neutral gene (NG). We applied this predictor to epigenetic regulator genes (ERGs), and used differential expression and correlation network analysis to identify dysregulated ERGs along with co-expressed cancer genes. Furthermore, we quantified global proteomic changes by mass spectrometry after EZH2 inhibition.
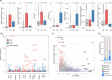
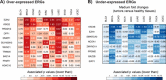
Results: Mutation-based classifiers uncovered the OG-like profile of DNMT3A and TSG-like profiles for several ERGs. Differential gene expression and correlation network analyses revealed that EZH2 is the most significantly over-expressed ERG in cancer and is co-regulated with a cell cycle network. Proteomic analysis showed that EZH2 inhibition induced down-regulation of cell cycle regulators in lymphoma cells.
Conclusions: Using classical driver genes to train an OG/TSG predictor, we determined the most predictive features at the gene level. Our predictor uncovered one OG and several TSGs among ERGs. Expression analyses elucidated multiple dysregulated ERGs including EZH2 as member of a co-expressed cell cycle network.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

