NET-GE: a novel NETwork-based Gene Enrichment for detecting biological processes associated to Mendelian diseases
- PMID: 26110971
- PMCID: PMC4480278
- DOI: 10.1186/1471-2164-16-S8-S6
NET-GE: a novel NETwork-based Gene Enrichment for detecting biological processes associated to Mendelian diseases
Abstract
Background: Enrichment analysis is a widely applied procedure for shedding light on the molecular mechanisms and functions at the basis of phenotypes, for enlarging the dataset of possibly related genes/proteins and for helping interpretation and prioritization of newly determined variations. Several standard and Network-based enrichment methods are available. Both approaches rely on the annotations that characterize the genes/proteins included in the input set; network based ones also include in different ways physical and functional relationships among different genes or proteins that can be extracted from the available biological networks of interactions.
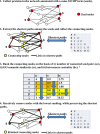
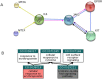
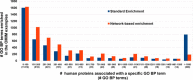
Results: Here we describe a novel procedure based on the extraction from the STRING interactome of sub-networks connecting proteins that share the same Gene Ontology(GO) terms for Biological Process (BP). Enrichment analysis is performed by mapping the protein set to be analyzed on the sub-networks, and then by collecting the corresponding annotations. We test the ability of our enrichment method in finding annotation terms disregarded by other enrichment methods available. We benchmarked 244 sets of proteins associated to different Mendelian diseases, according to the OMIM web resource. In 143 cases (58%), the network-based procedure extracts GO terms neglected by the standard method, and in 86 cases (35%), some of the newly enriched GO terms are not included in the set of annotations characterizing the input proteins. We present in detail six cases where our network-based enrichment provides an insight into the biological basis of the diseases, outperforming other freely available network-based methods.
Conclusions: Considering a set of proteins in the context of their interaction network can help in better defining their functions. Our novel method exploits the information contained in the STRING database for building the minimal connecting network containing all the proteins annotated with the same GO term. The enrichment procedure is performed considering the GO-specific network modules and, when tested on the OMIM-derived benchmark sets, it is able to extract enrichment terms neglected by other methods. Our procedure is effective even when the size of the input protein set is small, requiring at least two input proteins.
Figures
Similar articles
-
NET-GE: a web-server for NETwork-based human gene enrichment.Bioinformatics. 2016 Nov 15;32(22):3489-3491. doi: 10.1093/bioinformatics/btw508. Epub 2016 Aug 2. Bioinformatics. 2016. PMID: 27485441
-
eDGAR: a database of Disease-Gene Associations with annotated Relationships among genes.BMC Genomics. 2017 Aug 11;18(Suppl 5):554. doi: 10.1186/s12864-017-3911-3. BMC Genomics. 2017. PMID: 28812536 Free PMC article.
-
PhenPath: a tool for characterizing biological functions underlying different phenotypes.BMC Genomics. 2019 Jul 16;20(Suppl 8):548. doi: 10.1186/s12864-019-5868-x. BMC Genomics. 2019. PMID: 31307376 Free PMC article.
-
Discerning molecular interactions: A comprehensive review on biomolecular interaction databases and network analysis tools.Gene. 2018 Feb 5;642:84-94. doi: 10.1016/j.gene.2017.11.028. Epub 2017 Nov 10. Gene. 2018. PMID: 29129810 Review.
-
Exploring autophagy with Gene Ontology.Autophagy. 2018;14(3):419-436. doi: 10.1080/15548627.2017.1415189. Epub 2018 Feb 17. Autophagy. 2018. PMID: 29455577 Free PMC article. Review.
Cited by
-
Computational systems biology approaches for Parkinson's disease.Cell Tissue Res. 2018 Jul;373(1):91-109. doi: 10.1007/s00441-017-2734-5. Epub 2017 Nov 29. Cell Tissue Res. 2018. PMID: 29185073 Free PMC article. Review.
-
Overexpression of steroid sulfotransferase genes is associated with worsened prognosis and with immune exclusion in clear cell-renal cell carcinoma.Aging (Albany NY). 2019 Oct 27;11(20):9209-9219. doi: 10.18632/aging.102392. Epub 2019 Oct 27. Aging (Albany NY). 2019. PMID: 31655797 Free PMC article.
-
GCN-5/PGC-1α signaling is activated and associated with metabolism in cyclin E1-driven ovarian cancer.Aging (Albany NY). 2019 Jul 17;11(14):4890-4899. doi: 10.18632/aging.102082. Aging (Albany NY). 2019. PMID: 31313989 Free PMC article.
-
Knowledge-guided analysis of "omics" data using the KnowEnG cloud platform.PLoS Biol. 2020 Jan 23;18(1):e3000583. doi: 10.1371/journal.pbio.3000583. eCollection 2020 Jan. PLoS Biol. 2020. PMID: 31971940 Free PMC article.
-
An Overview of Two Old Friends Associated with Platelet Redox Signaling, the Protein Disulfide Isomerase and NADPH Oxidase.Biomolecules. 2023 May 17;13(5):848. doi: 10.3390/biom13050848. Biomolecules. 2023. PMID: 37238717 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

