AtRTD - a comprehensive reference transcript dataset resource for accurate quantification of transcript-specific expression in Arabidopsis thaliana
- PMID: 26111100
- PMCID: PMC4744958
- DOI: 10.1111/nph.13545
AtRTD - a comprehensive reference transcript dataset resource for accurate quantification of transcript-specific expression in Arabidopsis thaliana
Abstract
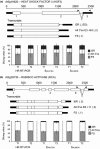
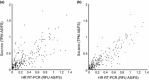
RNA-sequencing (RNA-seq) allows global gene expression analysis at the individual transcript level. Accurate quantification of transcript variants generated by alternative splicing (AS) remains a challenge. We have developed a comprehensive, nonredundant Arabidopsis reference transcript dataset (AtRTD) containing over 74 000 transcripts for use with algorithms to quantify AS transcript isoforms in RNA-seq. The AtRTD was formed by merging transcripts from TAIR10 and novel transcripts identified in an AS discovery project. We have estimated transcript abundance in RNA-seq data using the transcriptome-based alignment-free programmes Sailfish and Salmon and have validated quantification of splicing ratios from RNA-seq by high resolution reverse transcription polymerase chain reaction (HR RT-PCR). Good correlations between splicing ratios from RNA-seq and HR RT-PCR were obtained demonstrating the accuracy of abundances calculated for individual transcripts in RNA-seq. The AtRTD is a resource that will have immediate utility in analysing Arabidopsis RNA-seq data to quantify differential transcript abundance and expression.
Keywords: Arabidopsis thaliana; RNA-sequencing (RNA-seq); Sailfish; Salmon; alternative splicing; high resolution reverse transcription (HR RT)-PCR; transcripts per million.
© 2015 The Authors. New Phytologist © 2015 New Phytologist Trust.
Figures

References
-
- Airoldi CA, Davies B. 2012. Gene duplication and the evolution of plant MADS‐box transcription factors. Journal of Genetics and Genomics 39: 157–165. - PubMed
-
- Alamancos GP, Pagès A, Trincado JL, Bellora N, Eyras E. 2015. Leveraging transcript quantification for fast computation of alternative splicing profiles. bioRxiv. doi: 10.1101/008763. - DOI - PMC - PubMed
-
- Bardou F, Ariel F, Simpson CG, Romero‐Barrios N, Laporte P, Balzerque S, Brown JW, Crespi M. 2014. Long noncoding RNA modulates alternative splicing regulators in Arabidopsis . Developmental Cell 30: 166–176. - PubMed
-
- Black DL. 2003. Mechanisms of alternative pre‐messenger RNA splicing. Annual Review of Biochemistry 72: 291–336. - PubMed
-
- Carvalho RF, Feijão CV, Duque P. 2013. On the physiological significance of alternative splicing events in higher plants. Protoplasma 250: 639–650. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

