Integrating Image-Based Phenomics and Association Analysis to Dissect the Genetic Architecture of Temporal Salinity Responses in Rice
- PMID: 26111541
- PMCID: PMC4528749
- DOI: 10.1104/pp.15.00450
Integrating Image-Based Phenomics and Association Analysis to Dissect the Genetic Architecture of Temporal Salinity Responses in Rice
Abstract
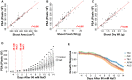
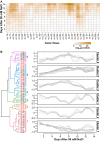
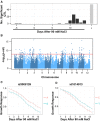
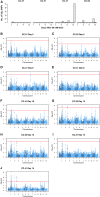
Salinity affects a significant portion of arable land and is particularly detrimental for irrigated agriculture, which provides one-third of the global food supply. Rice (Oryza sativa), the most important food crop, is salt sensitive. The genetic resources for salt tolerance in rice germplasm exist but are underutilized due to the difficulty in capturing the dynamic nature of physiological responses to salt stress. The genetic basis of these physiological responses is predicted to be polygenic. In an effort to address this challenge, we generated temporal imaging data from 378 diverse rice genotypes across 14 d of 90 mm NaCl stress and developed a statistical model to assess the genetic architecture of dynamic salinity-induced growth responses in rice germplasm. A genomic region on chromosome 3 was strongly associated with the early growth response and was captured using visible range imaging. Fluorescence imaging identified four genomic regions linked to salinity-induced fluorescence responses. A region on chromosome 1 regulates both the fluorescence shift indicative of the longer term ionic stress and the early growth rate decline during salinity stress. We present, to our knowledge, a new approach to capture the dynamic plant responses to its environment and elucidate the genetic basis of these responses using a longitudinal genome-wide association model.
© 2015 American Society of Plant Biologists. All Rights Reserved.
Figures
References
-
- Apse MP, Aharon GS, Snedden WA, Blumwald E (1999) Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285: 1256–1258 - PubMed
-
- Asano T, Hayashi N, Kobayashi M, Aoki N, Miyao A, Mitsuhara I, Ichikawa H, Komatsu S, Hirochika H, Kikuchi S, et al. (2012) A rice calcium-dependent protein kinase OsCPK12 oppositely modulates salt-stress tolerance and blast disease resistance. Plant J 69: 26–36 - PubMed
-
- Bac-Molenaar JA, Vreugdenhil D, Granier C, Keurentjes JJB (April 28, 2015) Genome-wide association mapping of growth dynamics detects time-specific and general quantitative trait loci. J Exp Bot http://dx.doi.org/10.1093/jxb/erv176 - DOI - PMC - PubMed
-
- Berger B, Parent B, Tester M (2010) High-throughput shoot imaging to study drought responses. J Exp Bot 61: 3519–3528 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

