The TatC component of the twin-arginine protein translocase functions as an obligate oligomer
- PMID: 26112072
- PMCID: PMC5102672
- DOI: 10.1111/mmi.13106
The TatC component of the twin-arginine protein translocase functions as an obligate oligomer
Abstract
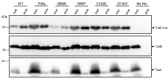
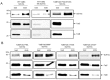
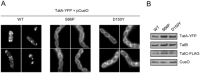
The Tat protein export system translocates folded proteins across the bacterial cytoplasmic membrane and the plant thylakoid membrane. The Tat system in Escherichia coli is composed of TatA, TatB and TatC proteins. TatB and TatC form an oligomeric, multivalent receptor complex that binds Tat substrates, while multiple protomers of TatA assemble at substrate-bound TatBC receptors to facilitate substrate transport. We have addressed whether oligomerisation of TatC is an absolute requirement for operation of the Tat pathway by screening for dominant negative alleles of tatC that inactivate Tat function in the presence of wild-type tatC. Single substitutions that confer dominant negative TatC activity were localised to the periplasmic cap region. The variant TatC proteins retained the ability to interact with TatB and with a Tat substrate but were unable to support the in vivo assembly of TatA complexes. Blue-native PAGE analysis showed that the variant TatC proteins produced smaller TatBC complexes than the wild-type TatC protein. The substitutions did not alter disulphide crosslinking to neighbouring TatC molecules from positions in the periplasmic cap but abolished a substrate-induced disulphide crosslink in transmembrane helix 5 of TatC. Our findings show that TatC functions as an obligate oligomer.
© 2015 The Authors. Molecular Microbiology published by John Wiley & Sons Ltd.
Figures








References
-
- Alami, M. , Luke, I. , Deitermann, S. , Eisner, G. , Koch, H.G. , Brunner, J. , and Muller, M. (2003) Differential interactions between a twin‐arginine signal peptide and its translocase in Escherichia coli . Mol Cell 12: 937–946. - PubMed
-
- Barrett, C.M. , Mangels, D. , and Robinson, C. (2005) Mutations in subunits of the Escherichia coli twin‐arginine translocase block function via differing effects on translocation activity or tat complex structure. J Mol Biol 347: 453–463. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

