Structural and Physical Basis for Anti-IgE Therapy
- PMID: 26113483
- PMCID: PMC4481376
- DOI: 10.1038/srep11581
Structural and Physical Basis for Anti-IgE Therapy
Abstract
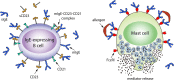
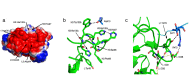
Omalizumab, an anti-IgE antibody, used to treat severe allergic asthma and chronic idiopathic urticaria, binds to IgE in blood or membrane-bound on B lymphocytes but not to IgE bound to its high (FcεRI) or low (CD23) affinity receptor. Mutagenesis studies indicate overlapping FcεRI and omalizumab-binding sites in the Cε3 domain, but crystallographic studies show FcεRI and CD23-binding sites that are far apart, so how can omalizumab block IgE from binding both receptors? We report a 2.42-Å omalizumab-Fab structure, a docked IgE-Fc/omalizumab-Fab structure consistent with available experimental data, and the free energy contributions of IgE residues to binding omalizumab, CD23, and FcεRI. These results provide a structural and physical basis as to why omalizumab cannot bind receptor-bound IgE and why omalizumab-bound IgE cannot bind to CD23/FcεRI. They reveal the key IgE residues and their roles in binding omalizumab, CD23, and FcεRI.
Figures





References
-
- Gould H. J. & Sutton B. J. IgE in allergy and asthma today. Nat. Rev. Immunol. 8, 205–217 (2008). - PubMed
-
- Chang T. W. et al.. Monoclonal antibodies specific for human IgE-producing B cells: a potential therapeutic for IgE-mediated allergic diseases. Bio/technology 8, 122–126 (1990). - PubMed
-
- Saini S. et al.. A randomized, placebo-controlled, dose-ranging study of single-dose omalizumab in patients with H1-antihistamine-refractory chronic idiopathic urticaria. J. Allergy Clin. Immunol. 128, 567–573 (2011). - PubMed
-
- Maurer M. et al.. Omalizumab for the treatment of chronic idiopathic or spontaneous urticaria. N. Engl. J. Med. 368, 924–935 (2013). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

